

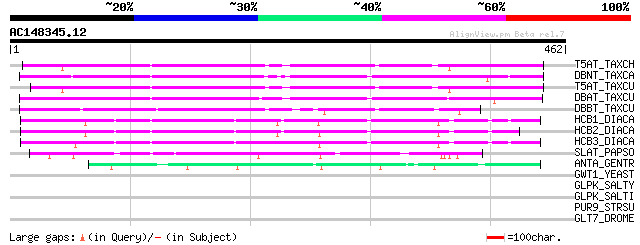
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148345.12 + phase: 0
(462 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
T5AT_TAXCH (Q8S9G6) Taxadien-5-alpha-ol O-acetyltransferase (EC ... 221 3e-57
DBNT_TAXCA (Q8LL69) 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransf... 218 3e-56
T5AT_TAXCU (Q9M6F0) Taxadien-5-alpha-ol O-acetyltransferase (EC ... 216 1e-55
DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III 10-O-acetyltransfera... 216 1e-55
DBBT_TAXCU (Q9FPW3) 2-alpha-hydroxytaxane 2-O-benzoyltransferase... 187 4e-47
HCB1_DIACA (O24645) Anthranilate N-benzoyltransferase protein 1 ... 154 6e-37
HCB2_DIACA (O23917) Anthranilate N-benzoyltransferase protein 2 ... 148 3e-35
HCB3_DIACA (O23918) Anthranilate N-benzoyltransferase protein 3 ... 147 6e-35
SLAT_PAPSO (Q94FT4) Salutaridinol 7-O-acetyltransferase (EC 2.3.... 80 1e-14
ANTA_GENTR (Q9ZWR8) Anthocyanin 5-aromatic acyltransferase (EC 2... 55 4e-07
GWT1_YEAST (P47026) GPI-anchored wall transfer protein 1 (EC 2.3... 32 3.4
GLPK_SALTY (Q8ZKP3) Glycerol kinase (EC 2.7.1.30) (ATP:glycerol ... 31 5.9
GLPK_SALTI (Q8Z2Y6) Glycerol kinase (EC 2.7.1.30) (ATP:glycerol ... 31 5.9
PUR9_STRSU (Q9F1T4) Bifunctional purine biosynthesis protein pur... 31 7.7
GLT7_DROME (Q8MV48) N-acetylgalactosaminyltransferase 7 (EC 2.4.... 31 7.7
>T5AT_TAXCH (Q8S9G6) Taxadien-5-alpha-ol O-acetyltransferase (EC
2.3.1.162)
(Taxa-4(20),11(12)-dien-5alpha-ol-O-acetyltransferase)
(Taxadienol acetyltransferase)
Length = 439
Score = 221 bits (563), Expect = 3e-57
Identities = 143/441 (32%), Positives = 225/441 (50%), Gaps = 23/441 (5%)
Query: 11 VRRSQPELVLPAAPTPHEVKLLSDIDDQEGLR---FNLPVIFVFRHEPSMKEKDPVKVLK 67
V ++ +V P+ P P LS ID+ G+R FN +I+ P+M DP K+++
Sbjct: 8 VNLNEKVMVGPSLPLPKTTLQLSSIDNLPGVRGSIFNALLIYNASPSPTMVSADPAKLIR 67
Query: 68 HALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQ 127
AL++ LVYY P AGR+RE L V+CTGEG MF+EA AD L GD P FQ
Sbjct: 68 EALAKILVYYPPFAGRLRETENGDLEVECTGEGAMFLEAMADNELSVLGD-FDDSNPSFQ 126
Query: 128 EILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGA 187
++L +P D P+ ++QVTR CGGF++ +S +H + DG G QF+ AE+ARG
Sbjct: 127 QLLFSLPLDTNFKDLPLLVVQVTRFTCGGFVVGVSFHHGVCDGRGAAQFLKGLAEMARGE 186
Query: 188 NQPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIA 247
+ S++P+WNRE++ DP Y+ H +E + P+ K +V F I
Sbjct: 187 VKLSLEPIWNRELVKLDDPKYLQFFH--FEFLRAPSIVEK------IVQTYFIIDFETIN 238
Query: 248 AIRRLVPLHLSR-CTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSL 306
I++ V C+++++ +A W RT+A Q+ E V+++ ++ R+ F+ S
Sbjct: 239 YIKQSVMEECKEFCSSFEVASAMTWIARTRAFQIPESEYVKILFGMDMRNSFNPPLPS-- 296
Query: 307 IGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCL-- 364
GYYGN I + AV ++L L A+ +I+K+K + + + V+K L
Sbjct: 297 -GYYGNSIGTACAVDNVQDLLSGSLLRAIMIIKKSKVSLNDNFKSR----AVVKPSELDV 351
Query: 365 -FTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLI 423
+DW+R F EV+FGWG AV + + FL + +G+
Sbjct: 352 NMNHENVVAFADWSRLGFDEVDFGWGNAVSVSPVQQQCELAMQNYFLFLKPSKNKPDGIK 411
Query: 424 LPICLPPEDMKRFVKELDDML 444
+ + LP MK F E++ M+
Sbjct: 412 ILMFLPLSKMKSFKIEMEAMM 432
>DBNT_TAXCA (Q8LL69) 3'-N-debenzoyl-2'-deoxytaxol
N-benzoyltransferase (EC 2.3.1.-) (DBTNBT)
Length = 441
Score = 218 bits (555), Expect = 3e-56
Identities = 143/440 (32%), Positives = 230/440 (51%), Gaps = 18/440 (4%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F V++ P +V P+ P+P LS +D R + VF + P DPVK+++
Sbjct: 9 FHVKKFDPVMVAPSLPSPKATVQLSVVDSLTICRGIFNTLLVF-NAPDNISADPVKIIRE 67
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQE 128
ALS+ LVYY+P AGR+R +L V+CTG+G +F+EA + T+ D L P FQ+
Sbjct: 68 ALSKVLVYYFPLAGRLRSKEIGELEVECTGDGALFVEAMVEDTISVLRD-LDDLNPSFQQ 126
Query: 129 ILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGAN 188
++ P I D + ++QVTR CGG + ++L H++ DG G QFV+A AE+ARG
Sbjct: 127 LVFWHPLDTAIEDLHLVIVQVTRFTCGGIAVGVTLPHSVCDGRGAAQFVTALAEMARGEV 186
Query: 189 QPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAA 248
+PS++P+WNRE+L DP ++ N +++ I PP + EE + SF I
Sbjct: 187 KPSLEPIWNRELLNPEDPLHLQLN--QFDSICPPP--MLEE----LGQASFVINVDTIEY 238
Query: 249 IRRLVPLHLSR-CTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
+++ V + C++++++ A W RTKALQ+ E+V+++ ++ R F N
Sbjct: 239 MKQCVMEECNEFCSSFEVVAALVWIARTKALQIPHTENVKLLFAMDLRKLF---NPPLPN 295
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
GYYGN I + A+ ++L L A+ +I+KAKA + + Y S
Sbjct: 296 GYYGNAIGTAYAMDNVQDLLNGSLLRAIMIIKKAKADLKDNYSRSRVVTNPYSLDVNKKS 355
Query: 368 GRTCMVSDWTRAKFSEVNFGWGEAVYGGA---AKGGIGSFLGGTFLVTHKNAKGEEGLIL 424
+SDW R F E +FGWG + + + G+ F +L+ KN K + +L
Sbjct: 356 DNILALSDWRRLGFYEADFGWGGPLNVSSLQRLENGLPMFSTFLYLLPAKN-KSDGIKLL 414
Query: 425 PICLPPEDMKRFVKELDDML 444
C+PP +K F ++ M+
Sbjct: 415 LSCMPPTTLKSFKIVMEAMI 434
>T5AT_TAXCU (Q9M6F0) Taxadien-5-alpha-ol O-acetyltransferase (EC
2.3.1.162)
(Taxa-4(20),11(12)-dien-5alpha-ol-O-acetyltransferase)
(Taxadienol acetyltransferase)
Length = 439
Score = 216 bits (550), Expect = 1e-55
Identities = 141/434 (32%), Positives = 220/434 (50%), Gaps = 23/434 (5%)
Query: 18 LVLPAAPTPHEVKLLSDIDDQEGLR---FNLPVIFVFRHEPSMKEKDPVKVLKHALSRTL 74
+V P+ P P LS ID+ G+R FN +I+ P+M DP K ++ AL++ L
Sbjct: 15 MVGPSPPLPKTTLQLSSIDNLPGVRGSIFNALLIYNASPSPTMISADPAKPIREALAKIL 74
Query: 75 VYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEILCDVP 134
VYY P AGR+RE L V+CTGEG MF+EA AD L GD P FQ++L +P
Sbjct: 75 VYYPPFAGRLRETENGDLEVECTGEGAMFLEAMADNELSVLGD-FDDSNPSFQQLLFSLP 133
Query: 135 GSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQPSIQP 194
D + ++QVTR CGGF++ +S +H + DG G QF+ AE+ARG + S++P
Sbjct: 134 LDTNFKDLSLLVVQVTRFTCGGFVVGVSFHHGVCDGRGAAQFLKGLAEMARGEVKLSLEP 193
Query: 195 VWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAAIRRLVP 254
+WNRE++ DP Y+ H +E + P+ K +V F I I++ V
Sbjct: 194 IWNRELVKLDDPKYLQFFH--FEFLRAPSIVEK------IVQTYFIIDFETINYIKQSVM 245
Query: 255 LHLSR-CTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLIGYYGNC 313
C+++++ +A W RT+A Q+ E V+++ ++ R+ F+ S GYYGN
Sbjct: 246 EECKEFCSSFEVASAMTWIARTRAFQIPESEYVKILFGMDMRNSFNPPLPS---GYYGNS 302
Query: 314 IAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCL---FTKGRT 370
I + AV ++L L A+ +I+K+K + + + V+K L
Sbjct: 303 IGTACAVDNVQDLLSGSLLRAIMIIKKSKVSLNDNFKSR----AVVKPSELDVNMNHENV 358
Query: 371 CMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPICLPP 430
+DW+R F EV+FGWG AV + + FL + +G+ + + LP
Sbjct: 359 VAFADWSRLGFDEVDFGWGNAVSVSPVQQQSALAMQNYFLFLKPSKNKPDGIKILMFLPL 418
Query: 431 EDMKRFVKELDDML 444
MK F E++ M+
Sbjct: 419 SKMKSFKIEMEAMM 432
>DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III 10-O-acetyltransferase
(EC 2.3.1.167) (DBAT)
Length = 440
Score = 216 bits (550), Expect = 1e-55
Identities = 141/438 (32%), Positives = 220/438 (50%), Gaps = 15/438 (3%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F VR + +V P+ P+P LS +D+ G+R N+ + + DP KV++
Sbjct: 7 FVVRSLERVMVAPSQPSPKAFLQLSTLDNLPGVRENIFNTLLVYNASDRVSVDPAKVIRQ 66
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQE 128
ALS+ LVYY P AGR+R+ L V+CTGEG +F+EA AD L GD L P ++
Sbjct: 67 ALSKVLVYYSPFAGRLRKKENGDLEVECTGEGALFVEAMADTDLSVLGD-LDDYSPSLEQ 125
Query: 129 ILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGAN 188
+L +P I D ++QVTR CGGF++ +S H + DG G QF+ A E+ARG
Sbjct: 126 LLFCLPPDTDIEDIHPLVVQVTRFTCGGFVVGVSFCHGICDGLGAGQFLIAMGEMARGEI 185
Query: 189 QPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAA 248
+PS +P+W RE+L DP Y + ++ I PP+T+ K +V S TS I
Sbjct: 186 KPSSEPIWKRELLKPEDPLY-RFQYYHFQLICPPSTFGK------IVQGSLVITSETINC 238
Query: 249 IRRLVPLHLSR-CTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSLI 307
I++ + C+A+++++A W RT+ALQ+ E+V+++ ++ R F N
Sbjct: 239 IKQCLREESKEFCSAFEVVSALAWIARTRALQIPHSENVKLIFAMDMRKLF---NPPLSK 295
Query: 308 GYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCLFTK 367
GYYGN + A+ K+L L V +I+KAK + E + ++ + +
Sbjct: 296 GYYGNFVGTVCAMDNVKDLLSGSLLRVVRIIKKAKVSLNEHFTSTIVTPRSGSDESI-NY 354
Query: 368 GRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIG--SFLGGTFLVTHKNAKGEEGLILP 425
D R F EV+FGWG A + G+ S + FL +G+ +
Sbjct: 355 ENIVGFGDRRRLGFDEVDFGWGHADNVSLVQHGLKDVSVVQSYFLFIRPPKNNPDGIKIL 414
Query: 426 ICLPPEDMKRFVKELDDM 443
+PP +K F E++ M
Sbjct: 415 SFMPPSIVKSFKFEMETM 432
>DBBT_TAXCU (Q9FPW3) 2-alpha-hydroxytaxane 2-O-benzoyltransferase
(EC 2.3.1.166) (TBT) (2-debenzoyl-7,13-diacetylbaccatin
III-2-O-benzoyl transferase) (DBBT)
Length = 440
Score = 187 bits (476), Expect = 4e-47
Identities = 122/391 (31%), Positives = 200/391 (50%), Gaps = 27/391 (6%)
Query: 9 FTVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVLKH 68
F V + +V P +P + LS ID++ N+ ++ S+ DP K ++
Sbjct: 4 FNVDMIERVIVAPCLQSPKNILHLSPIDNKTRGLTNILSVYNASQRVSVSA-DPAKTIRE 62
Query: 69 ALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQE 128
ALS+ LVYY P AGR+R L V+CTGEG +F+EA AD L D + P FQ+
Sbjct: 63 ALSKVLVYYPPFAGRLRNTENGDLEVECTGEGAVFVEAMADNDLSVLQDFNEYD-PSFQQ 121
Query: 129 ILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGAN 188
++ ++ I D + +QVTR CGGF++ +H++ DG G+ Q + E+ARG
Sbjct: 122 LVFNLREDVNIEDLHLLTVQVTRFTCGGFVVGTRFHHSVSDGKGIGQLLKGMGEMARGEF 181
Query: 189 QPSIQPVWNREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTSAHIAA 248
+PS++P+WNRE++ D Y+ +H ++ I PP K ++V+ + I
Sbjct: 182 KPSLEPIWNREMVKPEDIMYLQFDH--FDFIHPPLNLEKSIQASMVI------SFERINY 233
Query: 249 IRRLVPLHLSRC----TAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRS 304
I+R + + C +A++++ A W RTK+ ++ P+E V+++ ++ R+ F +
Sbjct: 234 IKRCM---MEECKEFFSAFEVVVALIWLARTKSFRIPPNEYVKIIFPIDMRNSFDSPLPK 290
Query: 305 SLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLADFMVIKERCL 364
GYYGN I + A+ K+L L YA+ LI+K+K + E + + + K L
Sbjct: 291 ---GYYGNAIGNACAMDNVKDLLNGSLLYALMLIKKSKFALNENFKSRI----LTKPSTL 343
Query: 365 FTKGRTCMV---SDWTRAKFSEVNFGWGEAV 392
+ V DW F E +FGWG AV
Sbjct: 344 DANMKHENVVGCGDWRNLGFYEADFGWGNAV 374
>HCB1_DIACA (O24645) Anthranilate N-benzoyltransferase protein 1 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 1)
Length = 445
Score = 154 bits (388), Expect = 6e-37
Identities = 121/458 (26%), Positives = 219/458 (47%), Gaps = 40/458 (8%)
Query: 10 TVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD-------- 61
+++ Q +V PA TP++ LS+ID ++ + +P E +
Sbjct: 2 SIQIKQSTMVRPAEETPNKSLWLSNIDMILRTPYSHTGAVLIYKQPDNNEDNIHPSSSMY 61
Query: 62 -PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQ 120
+L ALS+ LV +YP AGR++ G + +DC EG +F+EAE+ L+ FGD +
Sbjct: 62 FDANILIEALSKALVPFYPMAGRLKIN-GDRYEIDCNAEGALFVEAESSHVLEDFGD-FR 119
Query: 121 PPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAW 180
P + ++ S+ I P+ ++Q+TR +CGG + + +H + DG +F ++W
Sbjct: 120 PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHVCDGMAHFEFNNSW 179
Query: 181 AEIARGANQPSIQPVWNREI-LMARDPPYITCNHREYEQILP--PNTYIKEEDTTIVVHQ 237
A IA+G P+++PV +R + L R+PP I +H ++E +P PN + + T
Sbjct: 180 ARIAKGL-LPALEPVHDRYLHLRPRNPPQIKYSHSQFEPFVPSLPNELL--DGKTNKSQT 236
Query: 238 SFFFTSAHIAAIRRLVPL--HLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNAR 295
F + I +++ + L + +R + Y+++ A W +KA + E+++++ V+ R
Sbjct: 237 LFILSREQINTLKQKLDLSNNTTRLSTYEVVAAHVWRSVSKARGLSDHEEIKLIMPVDGR 296
Query: 296 SRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLAD 355
SR NN S GY GN + + T +L N L +++A + + Y+ S D
Sbjct: 297 SRI--NNPSLPKGYCGNVVFLAVCTATVGDLSCNPLTDTAGKVQEALKGLDDDYLRSAID 354
Query: 356 -----------FMVIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSF 404
+M E+ L+ +V+ W R + ++FGWG + G + +
Sbjct: 355 HTESKPGLPVPYMGSPEKTLYP---NVLVNSWGRIPYQAMDFGWGSPTFFGISN---IFY 408
Query: 405 LGGTFLVTHKNAKGEEGLILPICLPPEDMKRFVKELDD 442
G FL+ ++ G+ + L I L + RF K D
Sbjct: 409 DGQCFLIPSRD--GDGSMTLAINLFSSHLSRFKKYFYD 444
>HCB2_DIACA (O23917) Anthranilate N-benzoyltransferase protein 2 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 2)
Length = 446
Score = 148 bits (374), Expect = 3e-35
Identities = 116/441 (26%), Positives = 206/441 (46%), Gaps = 39/441 (8%)
Query: 10 TVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKD-------- 61
+++ Q +V PA TP++ LS ID ++ + +P E +
Sbjct: 2 SIQIKQSTMVRPAEETPNKSLWLSKIDMILRTPYSHTGAVLIYKQPDNNEDNIHPSSSMY 61
Query: 62 -PVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQ 120
+L ALS+ LV YYP AGR++ G + +DC EG +F+EAE+ L+ FGD +
Sbjct: 62 FDANILIEALSKALVPYYPMAGRLKIN-GDRYEIDCNAEGALFVEAESSHVLEDFGD-FR 119
Query: 121 PPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAW 180
P + ++ S+ I P+ ++Q+TR +CGG + + +H DG +F ++W
Sbjct: 120 PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHACDGMSHFEFNNSW 179
Query: 181 AEIARGANQPSIQPVWNREI-LMARDPPYITCNHREYEQILP--PNTYIKEEDTTIVVHQ 237
A IA+G P+++PV +R + L R+PP I H ++E +P PN + + T
Sbjct: 180 ARIAKGL-LPALEPVHDRYLHLRLRNPPQIKYTHSQFEPFVPSLPNELL--DGKTNKSQT 236
Query: 238 SFFFTSAHIAAIRRLVPLH---LSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNA 294
F + I +++ + L +R + Y+++ W +KA + E+++++ V+
Sbjct: 237 LFKLSREQINTLKQKLDLSSNTTTRLSTYEVVAGHVWRSVSKARGLSDHEEIKLIMPVDG 296
Query: 295 RSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLA 354
RSR NN S GY GN + + T +L N L +++A + + Y+ S
Sbjct: 297 RSRI--NNPSLPKGYCGNVVFLAVCTATVGDLSCNPLTDTAGKVQEALKGLDDDYLRSAI 354
Query: 355 D-----------FMVIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGS 403
D +M E+ L+ +V+ W R + ++FGWG + G +
Sbjct: 355 DHTESKPDLPVPYMGSPEKTLYP---NVLVNSWGRIPYQAMDFGWGSPTFFGISN---IF 408
Query: 404 FLGGTFLVTHKNAKGEEGLIL 424
+ G FL+ +N G L +
Sbjct: 409 YDGQCFLIPSQNGDGSMTLAI 429
>HCB3_DIACA (O23918) Anthranilate N-benzoyltransferase protein 3 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 3)
Length = 445
Score = 147 bits (371), Expect = 6e-35
Identities = 118/456 (25%), Positives = 211/456 (45%), Gaps = 36/456 (7%)
Query: 10 TVRRSQPELVLPAAPTPHEVKLLSDIDDQEGLRFN-LPVIFVFRH--------EPSMKEK 60
++ Q +V PA TP++ LS ID ++ + +++ +PS
Sbjct: 2 SIHIKQSTMVRPAEETPNKSLWLSKIDMILRTPYSHTGAVLIYKQPDNNEDNIQPSSSMY 61
Query: 61 DPVKVLKHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQ 120
+L ALS+ LV YYP AGR++ G + +DC GEG +F+EAE+ L+ FGD +
Sbjct: 62 FDANILIEALSKALVPYYPMAGRLKIN-GDRYEIDCNGEGALFVEAESSHVLEDFGD-FR 119
Query: 121 PPFPCFQEILCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAW 180
P + ++ S+ I P+ ++Q+TR +CGG + + +H + D +F ++W
Sbjct: 120 PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHVCDRMSHFEFNNSW 179
Query: 181 AEIARGANQPSIQPVWNREI-LMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSF 239
A IA+G P+++PV +R + L R+PP I H ++E +P + T F
Sbjct: 180 ARIAKGL-LPALEPVHDRYLHLCPRNPPQIKYTHSQFEPFVPSLPKELLDGKTSKSQTLF 238
Query: 240 FFTSAHIAAIRRLVPLH--LSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSR 297
+ I +++ + +R + Y+++ W +KA + E+++++ V+ RSR
Sbjct: 239 KLSREQINTLKQKLDWSNTTTRLSTYEVVAGHVWRSVSKARGLSDHEEIKLIMPVDGRSR 298
Query: 298 FSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLAD-- 355
NN S GY GN + + T +L N L +++A + + Y+ S D
Sbjct: 299 I--NNPSLPKGYCGNVVFLAVCTATVGDLACNPLTDTAGKVQEALKGLDDDYLRSAIDHT 356
Query: 356 ---------FMVIKERCLFTKGRTCMVSDWTRAKFSEVNFGWGEAVYGGAAKGGIGSFLG 406
+M E+ L+ +V+ W R + ++FGWG + G + + G
Sbjct: 357 ESKPDLPVPYMGSPEKTLYP---NVLVNSWGRIPYQAMDFGWGNPTFFGISN---IFYDG 410
Query: 407 GTFLVTHKNAKGEEGLILPICLPPEDMKRFVKELDD 442
FL+ +N G+ + L I L + F K D
Sbjct: 411 QCFLIPSQN--GDGSMTLAINLFSSHLSLFKKHFYD 444
>SLAT_PAPSO (Q94FT4) Salutaridinol 7-O-acetyltransferase (EC
2.3.1.150) (salAT)
Length = 474
Score = 79.7 bits (195), Expect = 1e-14
Identities = 92/427 (21%), Positives = 178/427 (41%), Gaps = 67/427 (15%)
Query: 17 ELVLPAAPTPHEVKL--LSDIDDQEGLRFNLPVIFVF-----RHEPSMKEKDPVKVLKHA 69
E + P PTP ++K LS +D L + +P+I + S D + +LK +
Sbjct: 15 ETIKPTTPTPSQLKNFNLSLLDQCFPLYYYVPIILFYPATAANSTGSSNHHDDLDLLKSS 74
Query: 70 LSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQPPFPCFQEI 129
LS+TLV++YP AGR+ + ++VDC +G+ F + + + +F QP P Q +
Sbjct: 75 LSKTLVHFYPMAGRMIDN----ILVDCHDQGINFYKVKIRGKMCEFMS--QPDVPLSQLL 128
Query: 130 LCDVPGSEYIIDRPIRLIQVTRLKCGGFILALSLNHTMGDGSGLRQFVSAWAEIARGANQ 189
+V + + + ++QV CGG + S++H + D + + F+ +WA + +
Sbjct: 129 PSEVVSAS-VPKEALVIVQVNMFDCGGTAICSSVSHKIADAATMSTFIRSWASTTKTSRS 187
Query: 190 -PSIQPVWNREILMARD-----PPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFFFTS 243
S V +++++ + D PP +P ++ ++ + V + F F
Sbjct: 188 GGSTAAVTDQKLIPSFDSASLFPPSERLTSPSGMSEIPFSSTPEDTEDDKTVSKRFVFDF 247
Query: 244 AHIAAIRRLVPLHL------SRCTAYDLITACYWCCRTKALQVEPDEDVRMM-CIVNARS 296
A I ++R + + + R T +++T+ W ++ P + ++ VN R
Sbjct: 248 AKITSVREKLQVLMHDNYKSRRQTRVEVVTSLIW---KSVMKSTPAGFLPVVHHAVNLRK 304
Query: 297 RFSANNRSSLIGYYGNCI-AFSAAVTTAKELCGNQLGYAVELIRKAKAQVTEKYMHSLAD 355
+ + G + AF A TT AV + + ++ +H L D
Sbjct: 305 KMDPPLQDVSFGNLSVTVSAFLPATTTTTT-------NAVNKTINSTSSESQVVLHELHD 357
Query: 356 FMV--------IK------ERCL--FTKGRTC-------------MVSDWTRAKFSEVNF 386
F+ +K E+ + F G +S W R E++F
Sbjct: 358 FIAQMRSEIDKVKGDKGSLEKVIQNFASGHDASIKKINDVEVINFWISSWCRMGLYEIDF 417
Query: 387 GWGEAVY 393
GWG+ ++
Sbjct: 418 GWGKPIW 424
>ANTA_GENTR (Q9ZWR8) Anthocyanin 5-aromatic acyltransferase (EC
2.3.1.153) (5AT)
Length = 469
Score = 55.1 bits (131), Expect = 4e-07
Identities = 97/415 (23%), Positives = 162/415 (38%), Gaps = 55/415 (13%)
Query: 66 LKHALSRTLVYYYPGAGR----IREGAGRKLMVDCT-GEGVMFIEAEADITLDQF-GDAL 119
LK +LS TL +Y P +G I+ G K G+ + I AE+D D G L
Sbjct: 66 LKASLSLTLKHYVPLSGNLLMPIKSGEMPKFQYSRDEGDSITLIVAESDQDFDYLKGHQL 125
Query: 120 QPPFPCFQEILCDVPGSEYIIDRPIRL-----------IQVTRLKCGGFILALSLNHTMG 168
D+ G Y++ R IR +QVT G +AL+ +H++
Sbjct: 126 VDSN--------DLHGLFYVMPRVIRTMQDYKVIPLVAVQVTVFPNRGIAVALTAHHSIA 177
Query: 169 DGSGLRQFVSAWAEIARGAN-----QPSIQPVWNREILMARDPPYITCNHREYEQILPPN 223
D F++AWA I + ++ P ++R I+ T + + + +
Sbjct: 178 DAKSFVMFINAWAYINKFGKDADLLSANLLPSFDRSIIKDLYGLEETFWNEMQDVLEMFS 237
Query: 224 TYIKEEDTTIVVHQSFFFTSAHIAAIRRLVPLHLS------RCTAYDLITACYWCCRTKA 277
+ + V ++ + A I ++ V L+L R T + + W C K+
Sbjct: 238 RFGSKPPRFNKVRATYVLSLAEIQKLKNKV-LNLRGSEPTIRVTTFTMTCGYVWTCMVKS 296
Query: 278 LQVEPDEDVRMMCIVNARSRFSANNRSSLI-----GYYGNCIAFSAAVTTAKELCGNQLG 332
E+ F+A+ R L Y+GNC+A A T KEL G++ G
Sbjct: 297 KDDVVSEESSNDENELEYFSFTADCRGLLTPPCPPNYFGNCLASCVAKATHKELVGDK-G 355
Query: 333 YAVELIRKAKAQVTEKYMHS----LADFMV-IKERCLFTKGRTCMVSDWTRAKFSEVNFG 387
V + A + EK +H+ LAD + E R ++ + V+FG
Sbjct: 356 LLVAV--AAIGEAIEKRLHNEKGVLADAKTWLSESNGIPSKRFLGITGSPKFDSYGVDFG 413
Query: 388 WGEAVYGGAAKGGIGSFLGGTFLVTHKNAKGEEGLILPICLPPEDMKRFVKELDD 442
WG+ AK I S + ++ E+G+ + + LP M F K ++
Sbjct: 414 WGK-----PAKFDITSVDYAELIYVIQSRDFEKGVEIGVSLPKIHMDAFAKIFEE 463
>GWT1_YEAST (P47026) GPI-anchored wall transfer protein 1 (EC
2.3.-.-) (Inositol acyltransferase GWT1)
Length = 498
Score = 32.0 bits (71), Expect = 3.4
Identities = 30/115 (26%), Positives = 47/115 (40%), Gaps = 17/115 (14%)
Query: 223 NTYIKEEDTTIVVHQSFFFTSA-------HIAAIRRLVPLHLSRCTAYDLITACYWCCRT 275
N +E T VH +FF T + I + R+VP RC+ I+ Y
Sbjct: 230 NLEYQEHVTEYGVHWNFFITLSLLPLVLTFIDPVTRMVP----RCSIAIFISCIY----- 280
Query: 276 KALQVEPDEDVRMMCIVNARSRFSANNRSSLIGYYGNCIAFSAAVTTAKELCGNQ 330
+ L ++ D + + + + FSAN R + + G C F T L GN+
Sbjct: 281 EWLLLKDDRTLNFLILADRNCFFSAN-REGIFSFLGYCSIFLWGQNTGFYLLGNK 334
>GLPK_SALTY (Q8ZKP3) Glycerol kinase (EC 2.7.1.30) (ATP:glycerol
3-phosphotransferase) (Glycerokinase) (GK)
Length = 501
Score = 31.2 bits (69), Expect = 5.9
Identities = 25/94 (26%), Positives = 40/94 (41%), Gaps = 13/94 (13%)
Query: 197 NREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFF---------FTSAHIA 247
+R ++M D ++ + RE+EQI P +++ + I QS +S IA
Sbjct: 16 SRAVVMDHDANIVSVSQREFEQIYPKPGWVEHDPMEIWASQSSTLVEVLAKADISSDQIA 75
Query: 248 AI----RRLVPLHLSRCTAYDLITACYWCCRTKA 277
AI +R + R T + A W CR A
Sbjct: 76 AIGITNQRETAIVWERETGKPIYNAIVWQCRRTA 109
>GLPK_SALTI (Q8Z2Y6) Glycerol kinase (EC 2.7.1.30) (ATP:glycerol
3-phosphotransferase) (Glycerokinase) (GK)
Length = 500
Score = 31.2 bits (69), Expect = 5.9
Identities = 25/94 (26%), Positives = 40/94 (41%), Gaps = 13/94 (13%)
Query: 197 NREILMARDPPYITCNHREYEQILPPNTYIKEEDTTIVVHQSFF---------FTSAHIA 247
+R ++M D ++ + RE+EQI P +++ + I QS +S IA
Sbjct: 15 SRAVVMDHDANIVSVSQREFEQIYPKPGWVEHDPMEIWASQSSTLVEVLAKADISSDQIA 74
Query: 248 AI----RRLVPLHLSRCTAYDLITACYWCCRTKA 277
AI +R + R T + A W CR A
Sbjct: 75 AIGITNQRETAIVWERETGKPIYNAIVWQCRRTA 108
>PUR9_STRSU (Q9F1T4) Bifunctional purine biosynthesis protein purH
[Includes: Phosphoribosylaminoimidazolecarboxamide
formyltransferase (EC 2.1.2.3) (AICAR transformylase);
IMP cyclohydrolase (EC 3.5.4.10) (Inosinicase) (IMP
synthetase) (ATIC)]
Length = 515
Score = 30.8 bits (68), Expect = 7.7
Identities = 26/108 (24%), Positives = 48/108 (44%), Gaps = 12/108 (11%)
Query: 247 AAIRRLVPLHLSRCTAYDLITACYWCCRTKALQVEPDEDVRMMCIVNARSRFSANNRSSL 306
A +RL AYD + A Y+ TK + + E + + +N R+ N + +
Sbjct: 166 ATRQRLAAKVFRHTAAYDALIADYF---TKQVGEDKPEKLTITYDLNQPMRYGENPQQNA 222
Query: 307 IGYYGNCIAFSAAVTTAKELCGNQLGY--------AVELIRKAKAQVT 346
+Y N + + ++ AK+L G +L + A+ +IR K + T
Sbjct: 223 -DFYQNALPTAYSIAAAKQLNGKELSFNNIRDADAAIRIIRDFKDRPT 269
>GLT7_DROME (Q8MV48) N-acetylgalactosaminyltransferase 7 (EC
2.4.1.-) (Protein-UDP acetylgalactosaminyltransferase 7)
(UDP-GalNAc:polypeptide
N-acetylgalactosaminyltransferase 7) (pp-GaNTase 7)
(dGalNAc-T2)
Length = 591
Score = 30.8 bits (68), Expect = 7.7
Identities = 29/115 (25%), Positives = 50/115 (43%), Gaps = 20/115 (17%)
Query: 9 FTVRRSQPELVLPAAPTP--HEVKLLSDIDDQEGLRFNLPVIFVFRHEPSMKEKDPVKVL 66
F+V V+ +PT HE+ L+ D D+E LR L E ++ K VKV+
Sbjct: 155 FSVLMRTVHSVIDRSPTHMLHEIILVDDFSDKENLRSQL-------DEYVLQFKGLVKVI 207
Query: 67 KHALSRTLVYYYPGAGRIREGAGRKLMVDCTGEGVMFIEAEADITLDQFGDALQP 121
++ L+ R R ++ TGE ++F++A ++ + L P
Sbjct: 208 RNKEREGLI-------RTRSRGA----MEATGEVIVFLDAHCEVNTNWLPPLLAP 251
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.139 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,757,072
Number of Sequences: 164201
Number of extensions: 2472604
Number of successful extensions: 5019
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 4970
Number of HSP's gapped (non-prelim): 17
length of query: 462
length of database: 59,974,054
effective HSP length: 114
effective length of query: 348
effective length of database: 41,255,140
effective search space: 14356788720
effective search space used: 14356788720
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 68 (30.8 bits)
Medicago: description of AC148345.12


