

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.3 + phase: 0 /pseudo
(456 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
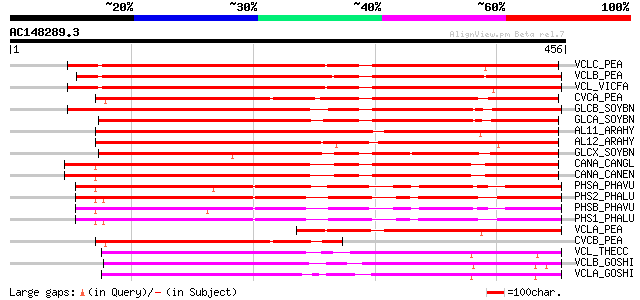
Score E
Sequences producing significant alignments: (bits) Value
VCLC_PEA (P13918) Vicilin precursor 597 e-170
VCLB_PEA (P02854) Provicilin precursor (Type B) (Fragment) 576 e-164
VCL_VICFA (P08438) Vicilin precursor 571 e-162
CVCA_PEA (P13915) Convicilin precursor 464 e-130
GLCB_SOYBN (P25974) Beta-conglycinin, beta chain precursor 421 e-117
GLCA_SOYBN (P13916) Beta-conglycinin, alpha chain precursor 388 e-107
AL11_ARAHY (P43237) Allergen Ara h 1, clone P17 precursor (Ara h I) 387 e-107
AL12_ARAHY (P43238) Allergen Ara h 1, clone P41B precursor (Ara ... 382 e-105
GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain precursor 376 e-104
CANA_CANGL (P10562) Canavalin precursor 363 e-100
CANA_CANEN (P50477) Canavalin precursor 363 e-100
PHSA_PHAVU (P07219) Phaseolin, alpha-type precursor 305 1e-82
PHS2_PHALU (Q43617) Phaseolin precursor (Fragment) 301 2e-81
PHSB_PHAVU (P02853) Phaseolin, beta-type precursor 299 1e-80
PHS1_PHALU (P80463) Phaseolin precursor 296 7e-80
VCLA_PEA (P02855) Provicilin (Type A) (Fragment) 291 3e-78
CVCB_PEA (P13919) Convicilin precursor (Fragment) 268 2e-71
VCL_THECC (Q43358) Vicilin precursor 253 5e-67
VCLB_GOSHI (P09801) Vicilin C72 precursor (Alpha-globulin B) 251 3e-66
VCLA_GOSHI (P09799) Vicilin GC72-A precursor (Alpha-globulin A) 246 9e-65
>VCLC_PEA (P13918) Vicilin precursor
Length = 459
Score = 597 bits (1540), Expect = e-170
Identities = 308/407 (75%), Positives = 353/407 (86%), Gaps = 16/407 (3%)
Query: 48 IKAPFQLLMLLGIFFLASVCVSSRSDHDQENPFFFNANRFQTLFENENGHIRLLQRFDKR 107
+KA F LLML+GI FLASVCVSSRSD +NPF F +N+FQTLFENENGHIRLLQ+FD+R
Sbjct: 6 MKASFPLLMLMGISFLASVCVSSRSD--PQNPFIFKSNKFQTLFENENGHIRLLQKFDQR 63
Query: 108 SKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGD 167
SKIFENLQNYRLLEY SKPHT+FLPQH DAD+IL VLSGKAILTVL P+DRNSFNLERGD
Sbjct: 64 SKIFENLQNYRLLEYKSKPHTIFLPQHTDADYILVVLSGKAILTVLKPDDRNSFNLERGD 123
Query: 168 TIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILE 227
TIKLPAG+IAYL NR DNE+LRVLDLAIPVNRPGQ QSF LSGNQNQQ++ SGFSKNILE
Sbjct: 124 TIKLPAGTIAYLVNRDDNEELRVLDLAIPVNRPGQLQSFLLSGNQNQQNYLSGFSKNILE 183
Query: 228 AAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSS 287
A+FN++YEEIE+VL+EE E+E +HRR L+D+R +QSQE NVIVK+SR QIEELS++AKS+
Sbjct: 184 ASFNTDYEEIEKVLLEEHEKETQHRRSLKDKR-QQSQEENVIVKLSRGQIEELSKNAKST 242
Query: 288 SRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAE 347
S++S SSES PFNLRSR PIYSNEFG FFEITPEKNPQLQDLDI VN E
Sbjct: 243 ----------SKKSVSSESEPFNLRSRGPIYSNEFGKFFEITPEKNPQLQDLDIFVNSVE 292
Query: 348 IREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQR---EYEEDEQEEERSQQV 404
I+EGSLLLPH+NSRA VIV V EGKG+FELVGQRNENQQEQR + EE++ EEE ++QV
Sbjct: 293 IKEGSLLLPHYNSRAIVIVTVNEGKGDFELVGQRNENQQEQRKEDDEEEEQGEEEINKQV 352
Query: 405 QRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLA 451
Q Y+A+L+ GDV+VIPAG+P VKASS+L LLGFGINAENNQR+FLA
Sbjct: 353 QNYKAKLSSGDVFVIPAGHPVAVKASSNLDLLGFGINAENNQRNFLA 399
Score = 34.3 bits (77), Expect = 0.68
Identities = 39/165 (23%), Positives = 69/165 (41%), Gaps = 27/165 (16%)
Query: 125 KPHTLFLPQHND-ADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGS------IA 177
K +L LP +N A I+ V GK ++ + N + D + G +
Sbjct: 294 KEGSLLLPHYNSRAIVIVTVNEGKGDFELVGQRNENQQEQRKEDDEEEEQGEEEINKQVQ 353
Query: 178 YLANRADNEDLRVLDLAIPVNRPGQFQ----SFSLSGNQNQQSFFSGFSKNIL------- 226
+ + D+ V+ PV F ++ NQ++F +G N++
Sbjct: 354 NYKAKLSSGDVFVIPAGHPVAVKASSNLDLLGFGINAENNQRNFLAGDEDNVISQIQRPV 413
Query: 227 -EAAFNSNYEEIERVLIEEQEQ------EPRHR-RGLRDRRHKQS 263
E AF + +E++R+L E Q+Q +P+ R RG R+ R + S
Sbjct: 414 KELAFPGSAQEVDRIL-ENQKQSHFADAQPQQRERGSRETRDRLS 457
>VCLB_PEA (P02854) Provicilin precursor (Type B) (Fragment)
Length = 410
Score = 576 bits (1484), Expect = e-164
Identities = 298/398 (74%), Positives = 341/398 (84%), Gaps = 14/398 (3%)
Query: 56 MLLGIFFLASVCVSSRSDHDQENPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQ 115
MLL I FLASVCVSSRSD QENPF F +NRFQTL+ENENGHIRLLQ+FDKRSKIFENLQ
Sbjct: 1 MLLAIAFLASVCVSSRSD--QENPFIFKSNRFQTLYENENGHIRLLQKFDKRSKIFENLQ 58
Query: 116 NYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGS 175
NYRLLEY SKPHTLFLPQ+ DADFIL VLSGKA LTVL NDRNSFNLERGD IKLPAGS
Sbjct: 59 NYRLLEYKSKPHTLFLPQYTDADFILVVLSGKATLTVLKSNDRNSFNLERGDAIKLPAGS 118
Query: 176 IAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYE 235
IAY ANR DNE+ RVLDLAIPVN+PGQ QSF LSG QNQ+S SGFSKNILEAAFN+NYE
Sbjct: 119 IAYFANRDDNEEPRVLDLAIPVNKPGQLQSFLLSGTQNQKSSLSGFSKNILEAAFNTNYE 178
Query: 236 EIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSE 295
EIE+VL+E+QEQEP+HRR L+DRR + ++E NVIVKVSR+QIEELS++AKSS
Sbjct: 179 EIEKVLLEQQEQEPQHRRSLKDRRQEINEE-NVIVKVSRDQIEELSKNAKSS-------- 229
Query: 296 SASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLL 355
S++S SSES PFNLRSR PIYSN+FG FFEITPEKN QLQDLDI VN +I+ GSLLL
Sbjct: 230 --SKKSVSSESGPFNLRSRNPIYSNKFGKFFEITPEKNQQLQDLDIFVNSVDIKVGSLLL 287
Query: 356 PHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGD 415
P++NSRA VIV V EGKG+FELVGQRNENQ ++ + +E+EQEEE S+QVQ YRA+L+PGD
Sbjct: 288 PNYNSRAIVIVTVTEGKGDFELVGQRNENQGKEND-KEEEQEEETSKQVQLYRAKLSPGD 346
Query: 416 VYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
V+VIPAG+P + ASSDL+L+G GINAENN+R+FLA +
Sbjct: 347 VFVIPAGHPVAINASSDLNLIGLGINAENNERNFLAGE 384
>VCL_VICFA (P08438) Vicilin precursor
Length = 463
Score = 571 bits (1471), Expect = e-162
Identities = 297/409 (72%), Positives = 344/409 (83%), Gaps = 16/409 (3%)
Query: 48 IKAPFQLLMLLGIFFLASVCVSSRSDHDQENPFFFNANRFQTLFENENGHIRLLQRFDKR 107
+K F LL LLGI FLASVC+SSRSD Q+NPF F +NRFQTLFENENGHIRLLQ+FD+
Sbjct: 6 LKDSFPLLTLLGIAFLASVCLSSRSD--QDNPFVFESNRFQTLFENENGHIRLLQKFDQH 63
Query: 108 SKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGD 167
SK+ ENLQNYRLLEY SKPHT+FLPQ DADFIL VLSGKAILTVL PNDRNSF+LERGD
Sbjct: 64 SKLLENLQNYRLLEYKSKPHTIFLPQQTDADFILVVLSGKAILTVLLPNDRNSFSLERGD 123
Query: 168 TIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILE 227
TIKLPAG+I YL NR D EDLRVLDL IPVNRPG+ QSF LSGNQNQ S SGFSKNILE
Sbjct: 124 TIKLPAGTIGYLVNRDDEEDLRVLDLVIPVNRPGEPQSFLLSGNQNQPSILSGFSKNILE 183
Query: 228 AAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSS 287
A+FN++Y+EIE+VL+EE +E HRRGL+DRR ++ QE NVIVK+SR+QIEEL+++AKSS
Sbjct: 184 ASFNTDYKEIEKVLLEEHGKEKYHRRGLKDRR-QRGQEENVIVKISRKQIEELNKNAKSS 242
Query: 288 SRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAE 347
S++S SSES PFNLRSREPIYSN+FG FFEITP++NPQLQDL+I VNY E
Sbjct: 243 ----------SKKSTSSESEPFNLRSREPIYSNKFGKFFEITPKRNPQLQDLNIFVNYVE 292
Query: 348 IREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQR-EYEEDEQ--EEERSQQV 404
I EGSLLLPH+NSRA VIV V EGKG+FELVGQRNENQQ R EY+E+++ EEE +QV
Sbjct: 293 INEGSLLLPHYNSRAIVIVTVNEGKGDFELVGQRNENQQGLREEYDEEKEQGEEEIRKQV 352
Query: 405 QRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
Q Y+A+L+PGDV VIPAGYP +KASS+L+L+GFGINAENNQR FLA +
Sbjct: 353 QNYKAKLSPGDVLVIPAGYPVAIKASSNLNLVGFGINAENNQRYFLAGE 401
>CVCA_PEA (P13915) Convicilin precursor
Length = 571
Score = 464 bits (1195), Expect = e-130
Identities = 243/385 (63%), Positives = 299/385 (77%), Gaps = 26/385 (6%)
Query: 71 RSDHDQE--NPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 128
RS QE NPF F +N+F TLFENENGHIR LQRFDKRS +FENLQNYRL+EY +KPHT
Sbjct: 144 RSSESQEHRNPFLFKSNKFLTLFENENGHIRRLQRFDKRSDLFENLQNYRLVEYRAKPHT 203
Query: 129 LFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDL 188
+FLPQH DAD IL VL+GKAILTVL+PNDRNS+NLERGDTIK+PAG+ +YL N+ D EDL
Sbjct: 204 IFLPQHIDADLILVVLNGKAILTVLSPNDRNSYNLERGDTIKIPAGTTSYLVNQDDEEDL 263
Query: 189 RVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQE 248
RV+D IPVNRPG+F++F LS N+NQ + GFSKNILEA+ N+ YE IE+VL+EEQE++
Sbjct: 264 RVVDFVIPVNRPGKFEAFGLSENKNQ--YLRGFSKNILEASLNTKYETIEKVLLEEQEKK 321
Query: 249 PRHRRGLRDR-RHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESA 307
P+ LRDR R +Q +E + I+KVSREQIEEL + AKSS S++S SE
Sbjct: 322 PQQ---LRDRKRTQQGEERDAIIKVSREQIEELRKLAKSS----------SKKSLPSEFE 368
Query: 308 PFNLRSREPIYSNEFGNFFEITPEKN-PQLQDLDILVNYAEIREGSLLLPHFNSRATVIV 366
PFNLRS +P YSN+FG FEITPEK PQLQDLDILV+ EI +G+L+LPH+NSRA V++
Sbjct: 369 PFNLRSHKPEYSNKFGKLFEITPEKKYPQLQDLDILVSCVEINKGALMLPHYNSRAIVVL 428
Query: 367 AVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNV 426
V EGKG EL+G +NE Q E E +ER+ +VQRY ARL+PGDV +IPAG+P
Sbjct: 429 LVNEGKGNLELLGLKNEQQ-------EREDRKERNNEVQRYEARLSPGDVVIIPAGHPVA 481
Query: 427 VKASSDLSLLGFGINAENNQRSFLA 451
+ ASS+L+LLGFGINA+NNQR+FL+
Sbjct: 482 ISASSNLNLLGFGINAKNNQRNFLS 506
Score = 32.3 bits (72), Expect = 2.6
Identities = 14/22 (63%), Positives = 18/22 (81%)
Query: 48 IKAPFQLLMLLGIFFLASVCVS 69
+K+ F LL+ LGI FLASVCV+
Sbjct: 5 VKSRFPLLLFLGIIFLASVCVT 26
Score = 32.0 bits (71), Expect = 3.4
Identities = 36/144 (25%), Positives = 54/144 (37%), Gaps = 42/144 (29%)
Query: 129 LFLPQHND-ADFILAVLSGKAILTVL----------------NPNDRNSFNLERGDTIKL 171
L LP +N A +L V GK L +L N R L GD + +
Sbjct: 415 LMLPHYNSRAIVVLLVNEGKGNLELLGLKNEQQEREDRKERNNEVQRYEARLSPGDVVII 474
Query: 172 PAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNIL----- 226
PAG +AI + F ++ NQ++F SG N++
Sbjct: 475 PAGH----------------PVAISASSNLNLLGFGINAKNNQRNFLSGSDDNVISQIEN 518
Query: 227 ---EAAFNSNYEEIERVLIEEQEQ 247
E F + +E+ R LI+ Q+Q
Sbjct: 519 PVKELTFPGSSQEVNR-LIKNQKQ 541
>GLCB_SOYBN (P25974) Beta-conglycinin, beta chain precursor
Length = 439
Score = 421 bits (1081), Expect = e-117
Identities = 222/407 (54%), Positives = 292/407 (71%), Gaps = 33/407 (8%)
Query: 48 IKAPFQLLMLLGIFFLASVCVSSRSDHDQENPFFF-NANRFQTLFENENGHIRLLQRFDK 106
++ F LL+LLG FLASVCVS + D+ NPF+F ++N FQTLFEN+N IRLLQRF+K
Sbjct: 2 MRVRFPLLVLLGTVFLASVCVSLKVREDENNPFYFRSSNSFQTLFENQNVRIRLLQRFNK 61
Query: 107 RSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERG 166
RS ENL++YR++++ SKP+T+ LP H DADF+L VLSG+AILT++N +DR+S+NL G
Sbjct: 62 RSPQLENLRDYRIVQFQSKPNTILLPHHADADFLLFVLSGRAILTLVNNDDRDSYNLHPG 121
Query: 167 DTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNIL 226
D ++PAG+ YL N D+++L+++ LAIPVN+PG++ F LS Q QQS+ GFS NIL
Sbjct: 122 DAQRIPAGTTYYLVNPHDHQNLKIIKLAIPVNKPGRYDDFFLSSTQAQQSYLQGFSHNIL 181
Query: 227 EAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKS 286
E +F+S +EEI RVL E+E+ Q Q+ VIV++S+EQI +LSR AKS
Sbjct: 182 ETSFHSEFEEINRVLFGEEEE--------------QRQQEGVIVELSKEQIRQLSRRAKS 227
Query: 287 SSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYA 346
S SR++ SSE PFNLRSR PIYSN FG FFEITPEKNPQL+DLDI ++
Sbjct: 228 S----------SRKTISSEDEPFNLRSRNPIYSNNFGKFFEITPEKNPQLRDLDIFLSSV 277
Query: 347 EIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQR 406
+I EG+LLLPHFNS+A VI+ + EG ELVG + E QQ+Q+ +EE +VQR
Sbjct: 278 DINEGALLLPHFNSKAIVILVINEGDANIELVGIK-EQQQKQK-------QEEEPLEVQR 329
Query: 407 YRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
YRA L+ DV+VIPA YP VV A+S+L+ L FGINAENNQR+FLA +
Sbjct: 330 YRAELSEDDVFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLAGE 376
>GLCA_SOYBN (P13916) Beta-conglycinin, alpha chain precursor
Length = 605
Score = 388 bits (996), Expect = e-107
Identities = 193/378 (51%), Positives = 279/378 (73%), Gaps = 27/378 (7%)
Query: 74 HDQENPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQ 133
H +NPF F +NRF+TLF+N+ G IR+LQRF++RS +NL++YR+LE++SKP+TL LP
Sbjct: 190 HKNKNPFLFGSNRFETLFKNQYGRIRVLQRFNQRSPQLQNLRDYRILEFNSKPNTLLLPN 249
Query: 134 HNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVLDL 193
H DAD+++ +L+G AIL+++N +DR+S+ L+ GD +++P+G+ Y+ N +NE+LR++ L
Sbjct: 250 HADADYLIVILNGTAILSLVNNDDRDSYRLQSGDALRVPSGTTYYVVNPDNNENLRLITL 309
Query: 194 AIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRR 253
AIPVN+PG+F+SF LS + QQS+ GFS+NILEA++++ +EEI +VL +E +
Sbjct: 310 AIPVNKPGRFESFFLSSTEAQQSYLQGFSRNILEASYDTKFEEINKVLFSREEGQ----- 364
Query: 254 GLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRS 313
++ +Q + +VIV++S+EQI LS+ AKSS SR++ SSE PFNLRS
Sbjct: 365 ----QQGEQRLQESVIVEISKEQIRALSKRAKSS----------SRKTISSEDKPFNLRS 410
Query: 314 REPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKG 373
R+PIYSN+ G FFEITPEKNPQL+DLDI ++ ++ EG+LLLPHFNS+A VI+ + EG
Sbjct: 411 RDPIYSNKLGKFFEITPEKNPQLRDLDIFLSIVDMNEGALLLPHFNSKAIVILVINEGDA 470
Query: 374 EFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKASSDL 433
ELVG + E QQEQ Q+EE+ +V++YRA L+ D++VIPAGYP VV A+S+L
Sbjct: 471 NIELVGLK-EQQQEQ-------QQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSNL 522
Query: 434 SLLGFGINAENNQRSFLA 451
+ GINAENNQR+FLA
Sbjct: 523 NFFAIGINAENNQRNFLA 540
>AL11_ARAHY (P43237) Allergen Ara h 1, clone P17 precursor (Ara h I)
Length = 614
Score = 387 bits (995), Expect = e-107
Identities = 194/388 (50%), Positives = 269/388 (69%), Gaps = 15/388 (3%)
Query: 71 RSDHDQENPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLF 130
R + + NPF+F + RF T + N+NG IR+LQRFD+RSK F+NLQN+R+++ ++P+TL
Sbjct: 160 REETSRNNPFYFPSRRFSTRYGNQNGRIRVLQRFDQRSKQFQNLQNHRIVQIEARPNTLV 219
Query: 131 LPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRV 190
LP+H DAD IL + G+A +TV N N+R SFNL+ G +++P+G I+Y+ NR DN++LRV
Sbjct: 220 LPKHADADNILVIQQGQATVTVANGNNRKSFNLDEGHALRIPSGFISYILNRHDNQNLRV 279
Query: 191 LDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPR 250
+++PVN PGQF+ F + +++Q S+ GFS+N LEAAFN+ + EI RVL+EE +
Sbjct: 280 AKISMPVNTPGQFEDFFPASSRDQSSYLQGFSRNTLEAAFNAEFNEIRRVLLEENAGGEQ 339
Query: 251 HRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFN 310
RG R R + S VIVKVS+E ++EL++HAKS S++ + E + P N
Sbjct: 340 EERGQRRRSTRSSDNEGVIVKVSKEHVQELTKHAKSVSKKGSEEEDITN--------PIN 391
Query: 311 LRSREPIYSNEFGNFFEITPE-KNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVE 369
LR EP SN FG FE+ P+ KNPQLQDLD+++ EI+EG+L+LPHFNS+A VIV V
Sbjct: 392 LRDGEPDLSNNFGRLFEVKPDKKNPQLQDLDMMLTCVEIKEGALMLPHFNSKAMVIVVVN 451
Query: 370 EGKGEFELVGQRNENQ------QEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGY 423
+G G ELV R E Q QE E EEDE+EE +++V+RY ARL GDV+++PA +
Sbjct: 452 KGTGNLELVAVRKEQQQRGRREQEWEEEEEDEEEEGSNREVRRYTARLKEGDVFIMPAAH 511
Query: 424 PNVVKASSDLSLLGFGINAENNQRSFLA 451
P + ASS+L LLGFGINAENN R FLA
Sbjct: 512 PVAINASSELHLLGFGINAENNHRIFLA 539
>AL12_ARAHY (P43238) Allergen Ara h 1, clone P41B precursor (Ara h
I)
Length = 626
Score = 382 bits (980), Expect = e-105
Identities = 198/387 (51%), Positives = 272/387 (70%), Gaps = 14/387 (3%)
Query: 71 RSDHDQENPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLF 130
R + + NPF+F + RF T + N+NG IR+LQRFD+RS+ F+NLQN+R+++ +KP+TL
Sbjct: 166 REETSRNNPFYFPSRRFSTRYGNQNGRIRVLQRFDQRSRQFQNLQNHRIVQIEAKPNTLV 225
Query: 131 LPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRV 190
LP+H DAD IL + G+A +TV N N+R SFNL+ G +++P+G I+Y+ NR DN++LRV
Sbjct: 226 LPKHADADNILVIQQGQATVTVANGNNRKSFNLDEGHALRIPSGFISYILNRHDNQNLRV 285
Query: 191 LDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPR 250
+++PVN PGQF+ F + +++Q S+ GFS+N LEAAFN+ + EI RVL+EE +
Sbjct: 286 AKISMPVNTPGQFEDFFPASSRDQSSYLQGFSRNTLEAAFNAEFNEIRRVLLEENAGGEQ 345
Query: 251 HRRGLRDRRHKQSQEAN--VIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAP 308
RG R R +S E N VIVKVS+E +EEL++HAKS S++ + E + P
Sbjct: 346 EERGQR-RWSTRSSENNEGVIVKVSKEHVEELTKHAKSVSKKGSEEE-------GDITNP 397
Query: 309 FNLRSREPIYSNEFGNFFEITPE-KNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVA 367
NLR EP SN FG FE+ P+ KNPQLQDLD+++ EI+EG+L+LPHFNS+A VIV
Sbjct: 398 INLREGEPDLSNNFGKLFEVKPDKKNPQLQDLDMMLTCVEIKEGALMLPHFNSKAMVIVV 457
Query: 368 VEEGKGEFELVGQRNENQQE-QREYEEDEQEEER--SQQVQRYRARLTPGDVYVIPAGYP 424
V +G G ELV R E QQ +RE EEDE EEE +++V+RY ARL GDV+++PA +P
Sbjct: 458 VNKGTGNLELVAVRKEQQQRGRREEEEDEDEEEEGSNREVRRYTARLKEGDVFIMPAAHP 517
Query: 425 NVVKASSDLSLLGFGINAENNQRSFLA 451
+ ASS+L LLGFGINAENN R FLA
Sbjct: 518 VAINASSELHLLGFGINAENNHRIFLA 544
>GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain precursor
Length = 639
Score = 376 bits (965), Expect = e-104
Identities = 191/397 (48%), Positives = 280/397 (70%), Gaps = 47/397 (11%)
Query: 74 HDQENPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQ 133
H +NPF FN+ RFQTLF+N+ GH+R+LQRF+KRS+ +NL++YR+LE++SKP+TL LP
Sbjct: 206 HKNKNPFHFNSKRFQTLFKNQYGHVRVLQRFNKRSQQLQNLRDYRILEFNSKPNTLLLPH 265
Query: 134 HNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLAN------------ 181
H DAD+++ +L+G AILT++N +DR+S+NL+ GD +++PAG+ Y+ N
Sbjct: 266 HADADYLIVILNGTAILTLVNNDDRDSYNLQSGDALRVPAGTTFYVVNPDNDENLRMIAG 325
Query: 182 ------RADN-EDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNY 234
DN E+LR++ LAIPVN+PG+F+SF LS Q QQS+ GFSKNILEA++++ +
Sbjct: 326 TTFYVVNPDNDENLRMITLAIPVNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKF 385
Query: 235 EEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASS 294
EEI +VL +E + + L++ +VIV++S++QI ELS+HAKSS
Sbjct: 386 EEINKVLFGREEGQQQGEERLQE---------SVIVEISKKQIRELSKHAKSS------- 429
Query: 295 ESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLL 354
SR++ SSE PFNL SR+PIYSN+ G FEIT ++NPQL+DLD+ ++ ++ EG+L
Sbjct: 430 ---SRKTISSEDKPFNLGSRDPIYSNKLGKLFEIT-QRNPQLRDLDVFLSVVDMNEGALF 485
Query: 355 LPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPG 414
LPHFNS+A V++ + EG+ ELVG + + Q+ +Q+EE+ +V++YRA L+
Sbjct: 486 LPHFNSKAIVVLVINEGEANIELVGIKEQQQR--------QQQEEQPLEVRKYRAELSEQ 537
Query: 415 DVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLA 451
D++VIPAGYP +V A+SDL+ FGINAENNQR+FLA
Sbjct: 538 DIFVIPAGYPVMVNATSDLNFFAFGINAENNQRNFLA 574
Score = 32.0 bits (71), Expect = 3.4
Identities = 33/152 (21%), Positives = 67/152 (43%), Gaps = 24/152 (15%)
Query: 129 LFLPQHNDADFILAVLS-GKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNED 187
LFLP N ++ V++ G+A + ++ ++ +R + P Y A ++ +D
Sbjct: 484 LFLPHFNSKAIVVLVINEGEANIELVGIKEQQ----QRQQQEEQPLEVRKYRAELSE-QD 538
Query: 188 LRVLDLAIPV----NRPGQFQSFSLSGNQNQQSFFSGFSKNIL--------EAAFNSNYE 235
+ V+ PV F +F ++ NQ++F +G N++ E AF + +
Sbjct: 539 IFVIPAGYPVMVNATSDLNFFAFGINAENNQRNFLAGSKDNVISQIPSQVQELAFPRSAK 598
Query: 236 EIERVLIEEQEQEPRHRRGLRDRRHKQSQEAN 267
+IE ++ + E D + +Q +E N
Sbjct: 599 DIENLIKSQSESY------FVDAQPQQKEEGN 624
>CANA_CANGL (P10562) Canavalin precursor
Length = 445
Score = 363 bits (933), Expect = e-100
Identities = 189/420 (45%), Positives = 278/420 (66%), Gaps = 54/420 (12%)
Query: 46 MAIKAPFQLLMLLGIFFLASVCVS--------------SRSDHDQENPFFFNANRFQTLF 91
MA A F L +LLG+ LASV S S Q NP+ F +N+F TLF
Sbjct: 1 MAFSARFPLWLLLGVVLLASVSASFAHSGHSGGEAEDESEESRAQNNPYLFRSNKFLTLF 60
Query: 92 ENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILT 151
+N++G +RLLQRF++ ++ ENL++YR+LEY SKP+TL LP H+D+D ++ VL G+AIL
Sbjct: 61 KNQHGSLRLLQRFNEDTEKLENLRDYRVLEYCSKPNTLLLPHHSDSDLLVLVLEGQAILV 120
Query: 152 VLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGN 211
++NP+ R+++ L++GD IK+ AG+ YL N +N++LR+L+ AI RPG + F LS
Sbjct: 121 LVNPDGRDTYKLDQGDAIKIQAGTPFYLINPDNNQNLRILNFAITFRRPGTVEDFFLSST 180
Query: 212 QNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVK 271
+ S+ S FSKN LEA+++S Y+EIE+ L++E+++ VIVK
Sbjct: 181 KRLPSYLSAFSKNFLEASYDSPYDEIEQTLLQEEQE-------------------GVIVK 221
Query: 272 VSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPE 331
+ ++QI+E+S+HA+SS SR++ SS+ PFNLRSR+PIYSN +G +EITPE
Sbjct: 222 MPKDQIQEISKHAQSS----------SRKTLSSQDKPFNLRSRDPIYSNNYGKLYEITPE 271
Query: 332 KNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREY 391
KN QL+DLDIL+N ++ EG+L +PH+NSRATVI+ EG+ E ELVG
Sbjct: 272 KNSQLRDLDILLNCLQMNEGALFVPHYNSRATVILVANEGRAEVELVG-----------L 320
Query: 392 EEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLA 451
E+ +Q+ S Q++RY A L+ GD+ VIP+ +P +KA+SDL+++G G+NAENN+R+FLA
Sbjct: 321 EQQQQQGLESMQLRRYAATLSEGDILVIPSSFPVALKAASDLNMVGIGVNAENNERNFLA 380
>CANA_CANEN (P50477) Canavalin precursor
Length = 445
Score = 363 bits (931), Expect = e-100
Identities = 189/420 (45%), Positives = 277/420 (65%), Gaps = 54/420 (12%)
Query: 46 MAIKAPFQLLMLLGIFFLASVCVS--------------SRSDHDQENPFFFNANRFQTLF 91
MA A F L +LLG+ LASV S S Q NP+ F +N+F TLF
Sbjct: 1 MAFSARFPLWLLLGVVLLASVSASFAHSGHSGGEAEDESEESRAQNNPYLFRSNKFLTLF 60
Query: 92 ENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILT 151
+N++G +RLLQRF++ ++ ENL++YR+LEY SKP+TL LP H+D+D ++ VL G+AIL
Sbjct: 61 KNQHGSLRLLQRFNEDTEKLENLRDYRVLEYCSKPNTLLLPHHSDSDLLVLVLEGQAILV 120
Query: 152 VLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGN 211
++NP+ R+++ L++GD IK+ AG+ YL N +N++LR+L AI RPG + F LS
Sbjct: 121 LVNPDGRDTYKLDQGDAIKIQAGTPFYLINPDNNQNLRILKFAITFRRPGTVEDFFLSST 180
Query: 212 QNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVK 271
+ S+ S FSKN LEA+++S Y+EIE+ L++E+++ VIVK
Sbjct: 181 KRLPSYLSAFSKNFLEASYDSPYDEIEQTLLQEEQE-------------------GVIVK 221
Query: 272 VSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPE 331
+ ++QI+E+S+HA+SS SR++ SS+ PFNLRSR+PIYSN +G +EITPE
Sbjct: 222 MPKDQIQEISKHAQSS----------SRKTLSSQDKPFNLRSRDPIYSNNYGKLYEITPE 271
Query: 332 KNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREY 391
KN QL+DLDIL+N ++ EG+L +PH+NSRATVI+ EG+ E ELVG
Sbjct: 272 KNSQLRDLDILLNCLQMNEGALFVPHYNSRATVILVANEGRAEVELVG-----------L 320
Query: 392 EEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLA 451
E+ +Q+ S Q++RY A L+ GD+ VIP+ +P +KA+SDL+++G G+NAENN+R+FLA
Sbjct: 321 EQQQQQGLESMQLRRYAATLSEGDIIVIPSSFPVALKAASDLNMVGIGVNAENNERNFLA 380
>PHSA_PHAVU (P07219) Phaseolin, alpha-type precursor
Length = 436
Score = 305 bits (782), Expect = 1e-82
Identities = 182/412 (44%), Positives = 248/412 (60%), Gaps = 66/412 (16%)
Query: 55 LMLLGIFFLASVCVS-----SRSDHDQENPFFFNA-NRFQTLFENENGHIRLLQRFDKRS 108
L+LLGI FLAS+ S + Q+NPF+FN+ N + TLF+N+ GHIR+LQRFD++S
Sbjct: 8 LLLLGILFLASLSASFATSLREEEESQDNPFYFNSDNSWNTLFKNQYGHIRVLQRFDQQS 67
Query: 109 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNS-FNLERG- 166
K +NL++YRL+E+ SKP TL LPQ DA+ +L V SG AIL ++ P+DR F L +G
Sbjct: 68 KRLQNLEDYRLVEFRSKPETLLLPQQADAELLLVVRSGSAILVLVKPDDRREYFFLTQGD 127
Query: 167 -----DTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGF 221
D K+PAG+I YL N EDLR++ LA+PVN P Q F LS + QQS+ F
Sbjct: 128 NPIFSDNQKIPAGTIFYLVNPDPKEDLRIIQLAMPVNNP-QIHEFFLSSTEAQQSYLQEF 186
Query: 222 SKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELS 281
SK+ILEA+FNS +EEI RVL EE+ Q+ ++ Q+ VIV + EQIEELS
Sbjct: 187 SKHILEASFNSKFEEINRVLFEEEGQQ------------EEGQQEGVIVNIDSEQIEELS 234
Query: 282 RHAKSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDI 341
+HAKSSSR+S S + + NEFGN E T L++
Sbjct: 235 KHAKSSSRKSHSKQ-------------------DNTIGNEFGNLTERTD------NSLNV 269
Query: 342 LVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERS 401
L++ E++EG+L +PH+ S+A VI+ V EG+ ELVG + +E E+E
Sbjct: 270 LISSIEMKEGALFVPHYYSKAIVILVVNEGEAHVELVGPK--GNKETLEFES-------- 319
Query: 402 QQVQRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
YRA L+ DV+VIPA YP +KA+S+++ GFGINA NN R+ LA K
Sbjct: 320 -----YRAELSKDDVFVIPAAYPVAIKATSNVNFTGFGINANNNNRNLLAGK 366
>PHS2_PHALU (Q43617) Phaseolin precursor (Fragment)
Length = 423
Score = 301 bits (772), Expect = 2e-81
Identities = 175/405 (43%), Positives = 248/405 (61%), Gaps = 64/405 (15%)
Query: 55 LMLLGIFFLASVCVS---SRSDHD--QENPFFFNA-NRFQTLFENENGHIRLLQRFDKRS 108
L+LLGI FLAS+ S S +H+ Q+NPF+F++ N +QTLF+N+ GHIR+LQRFD+RS
Sbjct: 5 LLLLGILFLASLSASFAISLREHNESQDNPFYFSSDNSWQTLFKNQYGHIRVLQRFDQRS 64
Query: 109 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDT 168
+ +NL++YRL+E+ SKP TL LPQ DA+F+L V SG A+L ++ P ++L++ DT
Sbjct: 65 ERLQNLEDYRLVEFMSKPETLLLPQQADAEFLLVVRSGSALLALVKPGGTIIYSLKQQDT 124
Query: 169 IKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEA 228
+K+PAG+I +L N +NEDLR++ LA+ VN P Q Q F LS + QQS+ GFSK+IL+A
Sbjct: 125 LKIPAGTIFFLINPENNEDLRIIKLAMTVNNP-QIQDFFLSSTEAQQSYLYGFSKHILDA 183
Query: 229 AFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSS 288
+FNS E+I R+L E + ++ VIV + + I+ELSRHAKSSS
Sbjct: 184 SFNSPIEKINRLLFAE-----------------EGRQEGVIVNIGSDLIQELSRHAKSSS 226
Query: 289 RRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEI 348
R+S S SNE+GN +I LD+L+ Y EI
Sbjct: 227 RKSLDHNSLD-------------------ISNEWGNLTDIV------YNSLDVLLTYVEI 261
Query: 349 REGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYR 408
+EG L +PH+NS+A VI+ VEEG + ELVG + E + S +++ YR
Sbjct: 262 KEGGLFVPHYNSKAIVILVVEEGVAKVELVGPKREKE---------------SLELETYR 306
Query: 409 ARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
A ++ GDV+VIPA +P +KA S+++ FGINA NN R FL K
Sbjct: 307 ADVSEGDVFVIPAAFPFAIKAISNVNFTSFGINANNNYRIFLTGK 351
>PHSB_PHAVU (P02853) Phaseolin, beta-type precursor
Length = 421
Score = 299 bits (765), Expect = 1e-80
Identities = 179/411 (43%), Positives = 241/411 (58%), Gaps = 70/411 (17%)
Query: 55 LMLLGIFFLASVCVS-----SRSDHDQENPFFFNA-NRFQTLFENENGHIRLLQRFDKRS 108
L+LLGI FLAS+ S + Q+NPF+FN+ N + TLF+N+ GHIR+LQRFD++S
Sbjct: 8 LLLLGILFLASLSASFATSLREEEESQDNPFYFNSDNSWNTLFKNQYGHIRVLQRFDQQS 67
Query: 109 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSF------N 162
K +NL++YRL+E+ SKP TL LPQ DA+ +L V SG AIL ++ P+DR + N
Sbjct: 68 KRLQNLEDYRLVEFRSKPETLLLPQQADAELLLVVRSGSAILVLVKPDDRREYFFLTSDN 127
Query: 163 LERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFS 222
D K+PAG+I YL N EDLR++ LA+PVN P Q F LS + QQS+ FS
Sbjct: 128 PIFSDHQKIPAGTIFYLVNPDPKEDLRIIQLAMPVNNP-QIHEFFLSSTEAQQSYLQEFS 186
Query: 223 KNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSR 282
K+ILEA+FNS +EEI RVL EE + Q+ VIV + EQI+ELS+
Sbjct: 187 KHILEASFNSKFEEINRVLFEE-----------------EGQQEGVIVNIDSEQIKELSK 229
Query: 283 HAKSSSRRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDIL 342
HAKSSSR+S S + + NEFGN E T L++L
Sbjct: 230 HAKSSSRKSLSKQ-------------------DNTIGNEFGNLTERTD------NSLNVL 264
Query: 343 VNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQ 402
++ E+ EG+L +PH+ S+A VI+ V EG+ ELVG + +E EYE
Sbjct: 265 ISSIEMEEGALFVPHYYSKAIVILVVNEGEAHVELVGPK--GNKETLEYES--------- 313
Query: 403 QVQRYRARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
YRA L+ DV+VIPA YP +KA+S+++ GFGINA NN R+ LA K
Sbjct: 314 ----YRAELSKDDVFVIPAAYPVAIKATSNVNFTGFGINANNNNRNLLAGK 360
>PHS1_PHALU (P80463) Phaseolin precursor
Length = 428
Score = 296 bits (758), Expect = 7e-80
Identities = 173/405 (42%), Positives = 244/405 (59%), Gaps = 64/405 (15%)
Query: 55 LMLLGIFFLASVCVS---SRSDHD--QENPFFFNA-NRFQTLFENENGHIRLLQRFDKRS 108
L+LLGI FLAS+ S S +H+ Q+NPF+F++ N +QTLF+N+ GHIR+LQ FD+ S
Sbjct: 8 LLLLGILFLASLSASFAISLREHNESQDNPFYFSSDNSWQTLFKNQYGHIRVLQSFDQHS 67
Query: 109 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDT 168
+ +NL++YRL+E+ SKP TL LPQ DA+F+L V SG A+L ++ P ++L++ DT
Sbjct: 68 ERLQNLEDYRLVEFMSKPETLLLPQQADAEFLLVVRSGSALLALVKPGGTIIYSLKQQDT 127
Query: 169 IKLPAGSIAYLANRADNEDLRVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEA 228
+K+PAG+I +L N +NEDLR++ LA+ VN P Q Q F LS + QQS+ GF K+IL+A
Sbjct: 128 LKIPAGTIFFLINPQNNEDLRIIKLAMTVNNP-QIQDFFLSSTEAQQSYLYGFRKDILDA 186
Query: 229 AFNSNYEEIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSS 288
+FNS EEI R+L E + ++ VIV + + I+ELSRHAKSSS
Sbjct: 187 SFNSPIEEINRLLFAE-----------------EGRQEGVIVNIGSDLIQELSRHAKSSS 229
Query: 289 RRSASSESASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEI 348
R+S S SNE+GN +I LD+L+ Y EI
Sbjct: 230 RKSLDHNSLD-------------------ISNEWGNLTDIV------YNSLDVLLTYVEI 264
Query: 349 REGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQEEERSQQVQRYR 408
+EG L +PH+NS+A VI+ VEEG + ELVG + E + S +++ YR
Sbjct: 265 KEGGLFVPHYNSKAIVILVVEEGVAKVELVGPKREKE---------------SLELETYR 309
Query: 409 ARLTPGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
A ++ GDV+VIPA YP +KA S+++ FGINA NN R L K
Sbjct: 310 ADVSEGDVFVIPAAYPVAIKAISNVNFTSFGINANNNYRILLTGK 354
>VCLA_PEA (P02855) Provicilin (Type A) (Fragment)
Length = 275
Score = 291 bits (744), Expect = 3e-78
Identities = 149/221 (67%), Positives = 183/221 (82%), Gaps = 14/221 (6%)
Query: 236 EIERVLIEEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSE 295
EIE++L+EE E+E HRRGLRD+R +QSQE NVIVKVS++QIEELS++AKSSS++S SS
Sbjct: 4 EIEKILLEEHEKETHHRRGLRDKR-QQSQEKNVIVKVSKKQIEELSKNAKSSSKKSVSSR 62
Query: 296 SASRRSASSESAPFNLRSREPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLL 355
S PFNL+S +PIYSN++G FFEITP+KNPQLQDLDI VNY EI+EGSL L
Sbjct: 63 SE----------PFNLKSSDPIYSNQYGKFFEITPKKNPQLQDLDIFVNYVEIKEGSLWL 112
Query: 356 PHFNSRATVIVAVEEGKGEFELVGQRNENQQ---EQREYEEDEQEEERSQQVQRYRARLT 412
PH+NSRA VIV V EGKG+FELVGQRNENQQ E+ + EE+++EEE QVQ Y+A+LT
Sbjct: 113 PHYNSRAIVIVTVNEGKGDFELVGQRNENQQGLREEDDEEEEQREEETKNQVQSYKAKLT 172
Query: 413 PGDVYVIPAGYPNVVKASSDLSLLGFGINAENNQRSFLAAK 453
PGDV+VIPAG+P V+ASS+L+LLGFGINAENNQR+FLA +
Sbjct: 173 PGDVFVIPAGHPVAVRASSNLNLLGFGINAENNQRNFLAGE 213
>CVCB_PEA (P13919) Convicilin precursor (Fragment)
Length = 386
Score = 268 bits (686), Expect = 2e-71
Identities = 135/205 (65%), Positives = 165/205 (79%), Gaps = 12/205 (5%)
Query: 71 RSDHDQE--NPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 128
RS QE NPF F +N+F TLFENENGHIRLLQRFDKRS +FENLQNYRL+EY +KPHT
Sbjct: 192 RSSESQERRNPFLFKSNKFLTLFENENGHIRLLQRFDKRSDLFENLQNYRLVEYRAKPHT 251
Query: 129 LFLPQHNDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDL 188
+FLPQH DAD IL VLSGKAILTVL+PNDRNS+NLERGDTIKLPAG+ +YL N+ D EDL
Sbjct: 252 IFLPQHIDADLILVVLSGKAILTVLSPNDRNSYNLERGDTIKLPAGTTSYLVNQDDEEDL 311
Query: 189 RVLDLAIPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQE 248
R++DL IPVN PG+F++F L+ N+NQ + GFSKNILEA++N+ YE IE+VL+EEQE++
Sbjct: 312 RLVDLVIPVNGPGKFEAFDLAKNKNQ--YLRGFSKNILEASYNTRYETIEKVLLEEQEKD 369
Query: 249 PRHRRGLRDRRHKQSQEANVIVKVS 273
+R +Q +E + IVKVS
Sbjct: 370 --------RKRRQQGEETDAIVKVS 386
Score = 31.2 bits (69), Expect = 5.8
Identities = 15/29 (51%), Positives = 22/29 (75%)
Query: 48 IKAPFQLLMLLGIFFLASVCVSSRSDHDQ 76
IK+ F LL+LLGI FLASV + +++D+
Sbjct: 5 IKSRFPLLLLLGIIFLASVVSVTYANYDE 33
>VCL_THECC (Q43358) Vicilin precursor
Length = 525
Score = 253 bits (647), Expect = 5e-67
Identities = 134/388 (34%), Positives = 223/388 (56%), Gaps = 34/388 (8%)
Query: 76 QENPFFFNANR-FQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQH 134
+ NP++F R FQT F +E G+ ++LQRF + S + + +YRL + + P+T LP H
Sbjct: 140 RNNPYYFPKRRSFQTRFRDEEGNFKILQRFAENSPPLKGINDYRLAMFEANPNTFILPHH 199
Query: 135 NDADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVLDLA 194
DA+ I V +GK +T + ++ S+N++RG + +PAGS Y+ ++ + E L + LA
Sbjct: 200 CDAEAIYFVTNGKGTITFVTHENKESYNVQRGTVVSVPAGSTVYVVSQDNQEKLTIAVLA 259
Query: 195 IPVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRG 254
+PVN PG+++ F +GN +S++ FS +LE FN+ E++E +L E+
Sbjct: 260 LPVNSPGKYELFFPAGNNKPESYYGAFSYEVLETVFNTQREKLEEILEEQ---------- 309
Query: 255 LRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSR 314
R ++ +Q Q+ + R AK R+ S ++ S R E NL S+
Sbjct: 310 -RGQKRQQGQQG-------------MFRKAKPEQIRAISQQATSPRHRGGERLAINLLSQ 355
Query: 315 EPIYSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGE 374
P+YSN+ G FFE PE Q Q++D+ V+ ++ +G++ +PH+NS+AT +V V +G G
Sbjct: 356 SPVYSNQNGRFFEACPEDFSQFQNMDVAVSAFKLNQGAIFVPHYNSKATFVVFVTDGYGY 415
Query: 375 FELV-------GQRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVV 427
++ Q +++ ++ R +E+E EEE + Q+ +A L+PGDV+V PAG+
Sbjct: 416 AQMACPHLSRQSQGSQSGRQDRREQEEESEEETFGEFQQVKAPLSPGDVFVAPAGHAVTF 475
Query: 428 KASSD--LSLLGFGINAENNQRSFLAAK 453
AS D L+ + FG+NA+NNQR FLA +
Sbjct: 476 FASKDQPLNAVAFGLNAQNNQRIFLAGR 503
>VCLB_GOSHI (P09801) Vicilin C72 precursor (Alpha-globulin B)
Length = 588
Score = 251 bits (641), Expect = 3e-66
Identities = 141/388 (36%), Positives = 219/388 (56%), Gaps = 37/388 (9%)
Query: 78 NPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDA 137
NPF F+ FQ+ F E+G+ R+LQRF R I + +RL + P+T LP H DA
Sbjct: 180 NPFHFHRRSFQSRFREEHGNFRVLQRFASRHPILRGINEFRLSILEANPNTFVLPHHCDA 239
Query: 138 DFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVLDLAIPV 197
+ I V +G+ LT L ++ S+N+ G +++PAGS YLAN+ + E L + L PV
Sbjct: 240 EKIYLVTNGRGTLTFLTHENKESYNVVPGVVVRVPAGSTVYLANQDNKEKLIIAVLHRPV 299
Query: 198 NRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGLRD 257
N P QF+ F +G+Q QS+ FS+ ILE AFN+ E+++ + Q
Sbjct: 300 NNPRQFEEFFPAGSQRPQSYLRAFSREILEPAFNTRSEQLDELFGGRQS----------- 348
Query: 258 RRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSREPI 317
H++ Q + K S+EQI R+ S E+ S R S E FNL R P
Sbjct: 349 --HRRQQGQGMFRKASQEQI------------RALSQEATSPREKSGERFAFNLLYRTPR 394
Query: 318 YSNEFGNFFEITPEKNPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFEL 377
YSN+ G F+E P + QL D+++ V+ ++ +GS+ +PH+NS+AT +V V EG G E+
Sbjct: 395 YSNQNGRFYEACPREFRQLSDINVTVSALQLNQGSIFVPHYNSKATFVVLVNEGNGYVEM 454
Query: 378 VG-----QRNENQQEQREYEEDEQEEERSQQVQRYRARLTPGDVYVIPAGYPNVVKAS-- 430
V Q + ++E+++ E++++EE RS Q ++ R++L+ GD++V+PA +P AS
Sbjct: 455 VSPHLPRQSSFEEEEEQQQEQEQEEERRSGQYRKIRSQLSRGDIFVVPANFPVTFVASQN 514
Query: 431 SDLSLLGFG-----INAENNQRSFLAAK 453
+L + GFG IN ++NQR F+A K
Sbjct: 515 QNLRMTGFGLYNQNINPDHNQRIFVAGK 542
Score = 33.5 bits (75), Expect = 1.2
Identities = 32/147 (21%), Positives = 59/147 (39%), Gaps = 7/147 (4%)
Query: 243 EEQEQEPRHRRGLRDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSA 302
+EQ+Q R + + H+Q Q + RE E + R + E +
Sbjct: 115 QEQQQSQRQFQECQQHCHQQEQRPERKQQCVRECRERYQENPWRREREEEAEEEETEEGE 174
Query: 303 SSES---APFNLRSREPIYSNEFGNF--FEITPEKNPQLQDL-DILVNYAEIREGSLLLP 356
+S F+ RS + + E GNF + ++P L+ + + ++ E + +LP
Sbjct: 175 QEQSHNPFHFHRRSFQSRFREEHGNFRVLQRFASRHPILRGINEFRLSILEANPNTFVLP 234
Query: 357 HFNSRATVIVAVEEGKGEFELVGQRNE 383
H + A I V G+G + N+
Sbjct: 235 H-HCDAEKIYLVTNGRGTLTFLTHENK 260
>VCLA_GOSHI (P09799) Vicilin GC72-A precursor (Alpha-globulin A)
Length = 605
Score = 246 bits (628), Expect = 9e-65
Identities = 144/387 (37%), Positives = 221/387 (56%), Gaps = 34/387 (8%)
Query: 76 QENPFFFNANRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHN 135
+ NP++F+ FQ F E+G+ R+LQRF + + + +R+ + P+T LP H
Sbjct: 179 RNNPYYFHRRSFQERFREEHGNFRVLQRFADKHHLLRGINEFRIAILEANPNTFVLPHHC 238
Query: 136 DADFILAVLSGKAILTVLNPNDRNSFNLERGDTIKLPAGSIAYLANRADNEDLRVLDLAI 195
DA+ I V +G+ +T + ++ S+N+ G +++PAGS YLAN+ + E L + L
Sbjct: 239 DAEKIYVVTNGRGTVTFVTHENKESYNVVPGVVVRIPAGSTVYLANQDNREKLTIAVLHR 298
Query: 196 PVNRPGQFQSFSLSGNQNQQSFFSGFSKNILEAAFNSNYEEIERVLIEEQEQEPRHRRGL 255
PVN PGQFQ F +G +N QS+ FS+ ILEA FN+ E+++ + P R+
Sbjct: 299 PVNNPGQFQKFFPAGQENPQSYLRIFSREILEAVFNTRSEQLDEL--------PGGRQS- 349
Query: 256 RDRRHKQSQEANVIVKVSREQIEELSRHAKSSSRRSASSESASRRSASSESAPFNLRSRE 315
H++ Q + K S+EQI LS+ A S R SE FNL S+
Sbjct: 350 ----HRRQQGQGMFRKASQEQIRALSQGA------------TSPRGKGSEGYAFNLLSQT 393
Query: 316 PIYSNEFGNFFEITPEK-NPQLQDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGE 374
P YSN+ G F+E P QL+++D V EI +GS+ +PH+NS+AT +V V EG G
Sbjct: 394 PRYSNQNGRFYEACPRNFQQQLREVDSSVVAFEINKGSIFVPHYNSKATFVVLVTEGNGH 453
Query: 375 FELV----GQRNENQQEQREYEEDEQE-EERSQQVQRYRARLTPGDVYVIPAGYPNVVKA 429
E+V +++ + + E E++EQE E RS Q +R RA+L+ G+++V+PAG+P A
Sbjct: 454 VEMVCPHLSRQSSDWSSREEEEQEEQEVERRSGQYKRVRAQLSTGNLFVVPAGHPVTFVA 513
Query: 430 S--SDLSLLGFGI-NAENNQRSFLAAK 453
S DL LLGFG+ N ++N+R F+A K
Sbjct: 514 SQNEDLGLLGFGLYNGQDNKRIFVAGK 540
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,746,110
Number of Sequences: 164201
Number of extensions: 2153083
Number of successful extensions: 14755
Number of sequences better than 10.0: 307
Number of HSP's better than 10.0 without gapping: 105
Number of HSP's successfully gapped in prelim test: 207
Number of HSP's that attempted gapping in prelim test: 12743
Number of HSP's gapped (non-prelim): 1222
length of query: 456
length of database: 59,974,054
effective HSP length: 114
effective length of query: 342
effective length of database: 41,255,140
effective search space: 14109257880
effective search space used: 14109257880
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148289.3


