

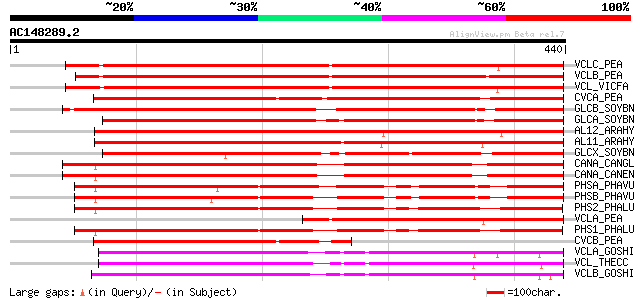
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148289.2 + phase: 0 /pseudo
(440 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VCLC_PEA (P13918) Vicilin precursor 602 e-172
VCLB_PEA (P02854) Provicilin precursor (Type B) (Fragment) 582 e-166
VCL_VICFA (P08438) Vicilin precursor 575 e-164
CVCA_PEA (P13915) Convicilin precursor 490 e-138
GLCB_SOYBN (P25974) Beta-conglycinin, beta chain precursor 424 e-118
GLCA_SOYBN (P13916) Beta-conglycinin, alpha chain precursor 400 e-111
AL12_ARAHY (P43238) Allergen Ara h 1, clone P41B precursor (Ara ... 397 e-110
AL11_ARAHY (P43237) Allergen Ara h 1, clone P17 precursor (Ara h I) 395 e-109
GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain precursor 382 e-106
CANA_CANGL (P10562) Canavalin precursor 372 e-103
CANA_CANEN (P50477) Canavalin precursor 372 e-102
PHSA_PHAVU (P07219) Phaseolin, alpha-type precursor 313 7e-85
PHSB_PHAVU (P02853) Phaseolin, beta-type precursor 305 2e-82
PHS2_PHALU (Q43617) Phaseolin precursor (Fragment) 301 3e-81
VCLA_PEA (P02855) Provicilin (Type A) (Fragment) 298 1e-80
PHS1_PHALU (P80463) Phaseolin precursor 295 1e-79
CVCB_PEA (P13919) Convicilin precursor (Fragment) 275 2e-73
VCLA_GOSHI (P09799) Vicilin GC72-A precursor (Alpha-globulin A) 259 7e-69
VCL_THECC (Q43358) Vicilin precursor 256 1e-67
VCLB_GOSHI (P09801) Vicilin C72 precursor (Alpha-globulin B) 253 7e-67
>VCLC_PEA (P13918) Vicilin precursor
Length = 459
Score = 602 bits (1551), Expect = e-172
Identities = 303/398 (76%), Positives = 356/398 (89%), Gaps = 7/398 (1%)
Query: 45 IKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKR 104
+KA LLML+GI FLAS+CVSSRSD +NPFIF SN+FQTLFENENGHIRLLQ+FD+R
Sbjct: 6 MKASFPLLMLMGISFLASVCVSSRSDP--QNPFIFKSNKFQTLFENENGHIRLLQKFDQR 63
Query: 105 SKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGD 164
SKIFENLQNYRLLEY SKPHT+FLPQH DAD+ILVV+SGKAILTVL P++RNSFNLERGD
Sbjct: 64 SKIFENLQNYRLLEYKSKPHTIFLPQHTDADYILVVLSGKAILTVLKPDDRNSFNLERGD 123
Query: 165 TIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILE 224
TIKLPAGT+ YL NRDDN++LRVLDLAIPVNRPGQ QSF LS ++NQQ++LSGFSKNILE
Sbjct: 124 TIKLPAGTIAYLVNRDDNEELRVLDLAIPVNRPGQLQSFLLSGNQNQQNYLSGFSKNILE 183
Query: 225 AAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKS 284
A+FN++YEEIE+VL+EE+E+E QHRR L+ ++RQQSQE NVIVK+SR QIEELSKNAKS
Sbjct: 184 ASFNTDYEEIEKVLLEEHEKETQHRRSLK--DKRQQSQEENVIVKLSRGQIEELSKNAKS 241
Query: 285 SSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLP 344
+S++S SSESEP NLR++ PIYSN+FG FFEITPEKNPQL+DLDI VN EI+EGSLLLP
Sbjct: 242 TSKKSVSSESEPFNLRSRGPIYSNEFGKFFEITPEKNPQLQDLDIFVNSVEIKEGSLLLP 301
Query: 345 HFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQ---ERSQQVQRYRARLSP 401
H+NSRA VIV V EGKG+FELVGQRNENQQEQR+ +++E++Q E ++QVQ Y+A+LS
Sbjct: 302 HYNSRAIVIVTVNEGKGDFELVGQRNENQQEQRKEDDEEEEQGEEEINKQVQNYKAKLSS 361
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
GDV+VIPAGHP+ V ASS+L LLGFGINAENNQRNFLA
Sbjct: 362 GDVFVIPAGHPVAVKASSNLDLLGFGINAENNQRNFLA 399
Score = 37.4 bits (85), Expect = 0.077
Identities = 48/222 (21%), Positives = 87/222 (38%), Gaps = 32/222 (14%)
Query: 66 SSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 125
S +S + PF S ++ NE G + + +++ ++L + + K +
Sbjct: 243 SKKSVSSESEPFNLRSRG--PIYSNEFG--KFFEITPEKNPQLQDLDIF-VNSVEIKEGS 297
Query: 126 LFLPQHNDADFILVVVS-GKAILTVLNPNNRNSFNLERGDTIKLPAG-------TLGYLA 177
L LP +N ++V V+ GK ++ N N + D + G Y A
Sbjct: 298 LLLPHYNSRAIVIVTVNEGKGDFELVGQRNENQQEQRKEDDEEEEQGEEEINKQVQNYKA 357
Query: 178 NRDDNKDLRVLDLAIPVNRPGQFQ----SFSLSESENQQSFLSGFSKNIL--------EA 225
D+ V+ PV F ++ NQ++FL+G N++ E
Sbjct: 358 KLSSG-DVFVIPAGHPVAVKASSNLDLLGFGINAENNQRNFLAGDEDNVISQIQRPVKEL 416
Query: 226 AFNSNYEEIERVLIEENE------QEPQHRRGLRKDERRQQS 261
AF + +E++R+L + + Q Q RG R+ R S
Sbjct: 417 AFPGSAQEVDRILENQKQSHFADAQPQQRERGSRETRDRLSS 458
>VCLB_PEA (P02854) Provicilin precursor (Type B) (Fragment)
Length = 410
Score = 582 bits (1499), Expect = e-166
Identities = 294/387 (75%), Positives = 341/387 (87%), Gaps = 5/387 (1%)
Query: 53 MLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQ 112
MLL I FLAS+CVSSRSDQ ENPFIF SNRFQTL+ENENGHIRLLQ+FDKRSKIFENLQ
Sbjct: 1 MLLAIAFLASVCVSSRSDQ--ENPFIFKSNRFQTLYENENGHIRLLQKFDKRSKIFENLQ 58
Query: 113 NYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGT 172
NYRLLEY SKPHTLFLPQ+ DADFILVV+SGKA LTVL N+RNSFNLERGD IKLPAG+
Sbjct: 59 NYRLLEYKSKPHTLFLPQYTDADFILVVLSGKATLTVLKSNDRNSFNLERGDAIKLPAGS 118
Query: 173 LGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYE 232
+ Y ANRDDN++ RVLDLAIPVN+PGQ QSF LS ++NQ+S LSGFSKNILEAAFN+NYE
Sbjct: 119 IAYFANRDDNEEPRVLDLAIPVNKPGQLQSFLLSGTQNQKSSLSGFSKNILEAAFNTNYE 178
Query: 233 EIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESS 292
EIE+VL+E+ EQEPQHRR L+ +RRQ+ E NVIVKVSR+QIEELSKNAKSSS++S SS
Sbjct: 179 EIEKVLLEQQEQEPQHRRSLK--DRRQEINEENVIVKVSRDQIEELSKNAKSSSKKSVSS 236
Query: 293 ESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATV 352
ES P NLR++ PIYSNKFG FFEITPEKN QL+DLDI VN +I+ GSLLLP++NSRA V
Sbjct: 237 ESGPFNLRSRNPIYSNKFGKFFEITPEKNQQLQDLDIFVNSVDIKVGSLLLPNYNSRAIV 296
Query: 353 IVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHP 412
IV V EGKG+FELVGQRNENQ ++ + +E+EQ++E S+QVQ YRA+LSPGDV+VIPAGHP
Sbjct: 297 IVTVTEGKGDFELVGQRNENQGKEND-KEEEQEEETSKQVQLYRAKLSPGDVFVIPAGHP 355
Query: 413 IVVTASSDLSLLGFGINAENNQRNFLA 439
+ + ASSDL+L+G GINAENN+RNFLA
Sbjct: 356 VAINASSDLNLIGLGINAENNERNFLA 382
>VCL_VICFA (P08438) Vicilin precursor
Length = 463
Score = 575 bits (1483), Expect = e-164
Identities = 292/398 (73%), Positives = 347/398 (86%), Gaps = 7/398 (1%)
Query: 45 IKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKR 104
+K LL LLGI FLAS+C+SSRSDQD NPF+F SNRFQTLFENENGHIRLLQ+FD+
Sbjct: 6 LKDSFPLLTLLGIAFLASVCLSSRSDQD--NPFVFESNRFQTLFENENGHIRLLQKFDQH 63
Query: 105 SKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGD 164
SK+ ENLQNYRLLEY SKPHT+FLPQ DADFILVV+SGKAILTVL PN+RNSF+LERGD
Sbjct: 64 SKLLENLQNYRLLEYKSKPHTIFLPQQTDADFILVVLSGKAILTVLLPNDRNSFSLERGD 123
Query: 165 TIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILE 224
TIKLPAGT+GYL NRDD +DLRVLDL IPVNRPG+ QSF LS ++NQ S LSGFSKNILE
Sbjct: 124 TIKLPAGTIGYLVNRDDEEDLRVLDLVIPVNRPGEPQSFLLSGNQNQPSILSGFSKNILE 183
Query: 225 AAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKS 284
A+FN++Y+EIE+VL+EE+ +E HRRGL+ +RRQ+ QE NVIVK+SR+QIEEL+KNAKS
Sbjct: 184 ASFNTDYKEIEKVLLEEHGKEKYHRRGLK--DRRQRGQEENVIVKISRKQIEELNKNAKS 241
Query: 285 SSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLP 344
SS++S SSESEP NLR+++PIYSNKFG FFEITP++NPQL+DL+I VNY EI EGSLLLP
Sbjct: 242 SSKKSTSSESEPFNLRSREPIYSNKFGKFFEITPKRNPQLQDLNIFVNYVEINEGSLLLP 301
Query: 345 HFNSRATVIVAVEEGKGEFELVGQRNENQQEQR-EYEEDEQQ--QERSQQVQRYRARLSP 401
H+NSRA VIV V EGKG+FELVGQRNENQQ R EY+E+++Q +E +QVQ Y+A+LSP
Sbjct: 302 HYNSRAIVIVTVNEGKGDFELVGQRNENQQGLREEYDEEKEQGEEEIRKQVQNYKAKLSP 361
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
GDV VIPAG+P+ + ASS+L+L+GFGINAENNQR FLA
Sbjct: 362 GDVLVIPAGYPVAIKASSNLNLVGFGINAENNQRYFLA 399
Score = 37.0 bits (84), Expect = 0.10
Identities = 48/218 (22%), Positives = 88/218 (40%), Gaps = 29/218 (13%)
Query: 66 SSRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHT 125
S +S + PF S + ++ N+ G + + KR+ ++L N + +
Sbjct: 243 SKKSTSSESEPFNLRSR--EPIYSNKFG--KFFEITPKRNPQLQDL-NIFVNYVEINEGS 297
Query: 126 LFLPQHNDADFILVVVS-GKAILTVLNPNNRNSFNL--------ERGDTIKLPAGTLGYL 176
L LP +N ++V V+ GK ++ N N L E+G+ ++ Y
Sbjct: 298 LLLPHYNSRAIVIVTVNEGKGDFELVGQRNENQQGLREEYDEEKEQGEE-EIRKQVQNYK 356
Query: 177 ANRDDNKDLRV---LDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL--------EA 225
A L + +AI + F ++ NQ+ FL+G N++ E
Sbjct: 357 AKLSPGDVLVIPAGYPVAIKASSNLNLVGFGINAENNQRYFLAGEEDNVISQIHKPVKEL 416
Query: 226 AFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQE 263
AF + +E++ +L Q+ H + ER + SQE
Sbjct: 417 AFPGSAQEVDTLL---ENQKQSHFANAQPRERERGSQE 451
>CVCA_PEA (P13915) Convicilin precursor
Length = 571
Score = 490 bits (1261), Expect = e-138
Identities = 246/374 (65%), Positives = 303/374 (80%), Gaps = 13/374 (3%)
Query: 67 SRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTL 126
S Q+ NPF+F SN+F TLFENENGHIR LQRFDKRS +FENLQNYRL+EY +KPHT+
Sbjct: 145 SSESQEHRNPFLFKSNKFLTLFENENGHIRRLQRFDKRSDLFENLQNYRLVEYRAKPHTI 204
Query: 127 FLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLR 186
FLPQH DAD ILVV++GKAILTVL+PN+RNS+NLERGDTIK+PAGT YL N+DD +DLR
Sbjct: 205 FLPQHIDADLILVVLNGKAILTVLSPNDRNSYNLERGDTIKIPAGTTSYLVNQDDEEDLR 264
Query: 187 VLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
V+D IPVNRPG+F++F LSE++NQ +L GFSKNILEA+ N+ YE IE+VL+EE E++P
Sbjct: 265 VVDFVIPVNRPGKFEAFGLSENKNQ--YLRGFSKNILEASLNTKYETIEKVLLEEQEKKP 322
Query: 247 QHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIY 306
Q LR +R QQ +E + I+KVSREQIEEL K AKSSS++S SE EP NLR+ KP Y
Sbjct: 323 QQ---LRDRKRTQQGEERDAIIKVSREQIEELRKLAKSSSKKSLPSEFEPFNLRSHKPEY 379
Query: 307 SNKFGNFFEITPEKN-PQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFEL 365
SNKFG FEITPEK PQL+DLDILV+ EI +G+L+LPH+NSRA V++ V EGKG EL
Sbjct: 380 SNKFGKLFEITPEKKYPQLQDLDILVSCVEINKGALMLPHYNSRAIVVLLVNEGKGNLEL 439
Query: 366 VGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLG 425
+G +NE Q E E ++ER+ +VQRY ARLSPGDV +IPAGHP+ ++ASS+L+LLG
Sbjct: 440 LGLKNEQQ-------EREDRKERNNEVQRYEARLSPGDVVIIPAGHPVAISASSNLNLLG 492
Query: 426 FGINAENNQRNFLA 439
FGINA+NNQRNFL+
Sbjct: 493 FGINAKNNQRNFLS 506
Score = 36.6 bits (83), Expect = 0.13
Identities = 37/162 (22%), Positives = 62/162 (37%), Gaps = 44/162 (27%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNN----------------RNSFNLERGDTIKL 168
L LP +N A +L+V GK L +L N R L GD + +
Sbjct: 415 LMLPHYNSRAIVVLLVNEGKGNLELLGLKNEQQEREDRKERNNEVQRYEARLSPGDVVII 474
Query: 169 PAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNIL----- 223
PAG +AI + F ++ NQ++FLSG N++
Sbjct: 475 PAGH----------------PVAISASSNLNLLGFGINAKNNQRNFLSGSDDNVISQIEN 518
Query: 224 ---EAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQ 262
E F + +E+ R++ Q+ H +++ ++SQ
Sbjct: 519 PVKELTFPGSSQEVNRLI---KNQKQSHFASAEPEQKEEESQ 557
>GLCB_SOYBN (P25974) Beta-conglycinin, beta chain precursor
Length = 439
Score = 424 bits (1091), Expect = e-118
Identities = 221/398 (55%), Positives = 294/398 (73%), Gaps = 26/398 (6%)
Query: 43 MAIKAPCSLLMLLGIVFLASICVSSRSDQDQENPFIF-NSNRFQTLFENENGHIRLLQRF 101
M ++ P LL+LLG VFLAS+CVS + +D+ NPF F +SN FQTLFEN+N IRLLQRF
Sbjct: 2 MRVRFP--LLVLLGTVFLASVCVSLKVREDENNPFYFRSSNSFQTLFENQNVRIRLLQRF 59
Query: 102 DKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLE 161
+KRS ENL++YR++++ SKP+T+ LP H DADF+L V+SG+AILT++N ++R+S+NL
Sbjct: 60 NKRSPQLENLRDYRIVQFQSKPNTILLPHHADADFLLFVLSGRAILTLVNNDDRDSYNLH 119
Query: 162 RGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKN 221
GD ++PAGT YL N D+++L+++ LAIPVN+PG++ F LS ++ QQS+L GFS N
Sbjct: 120 PGDAQRIPAGTTYYLVNPHDHQNLKIIKLAIPVNKPGRYDDFFLSSTQAQQSYLQGFSHN 179
Query: 222 ILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKN 281
ILE +F+S +EEI RVL E E +Q Q+ VIV++S+EQI +LS+
Sbjct: 180 ILETSFHSEFEEINRVLFGEEE---------------EQRQQEGVIVELSKEQIRQLSRR 224
Query: 282 AKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSL 341
AKSSSR++ SSE EP NLR++ PIYSN FG FFEITPEKNPQL+DLDI ++ +I EG+L
Sbjct: 225 AKSSSRKTISSEDEPFNLRSRNPIYSNNFGKFFEITPEKNPQLRDLDIFLSSVDINEGAL 284
Query: 342 LLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSP 401
LLPHFNS+A VI+ + EG ELVG + E QQ+Q+ Q+E +VQRYRA LS
Sbjct: 285 LLPHFNSKAIVILVINEGDANIELVGIK-EQQQKQK-------QEEEPLEVQRYRAELSE 336
Query: 402 GDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
DV+VIPA +P VV A+S+L+ L FGINAENNQRNFLA
Sbjct: 337 DDVFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLA 374
Score = 33.9 bits (76), Expect = 0.85
Identities = 33/131 (25%), Positives = 62/131 (47%), Gaps = 18/131 (13%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
L LP N A ILV+ G A + ++ + + + +++ Y A ++ D
Sbjct: 284 LLLPHFNSKAIVILVINEGDANIELVGIKEQQQKQKQEEEPLEVQR----YRAELSED-D 338
Query: 185 LRVLDLAIP--VNRPGQ--FQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ A P VN F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 339 VFVIPAAYPFVVNATSNLNFLAFGINAENNQRNFLAGEKDNVVRQIERQVQELAFPGSAQ 398
Query: 233 EIERVLIEENE 243
++ER+L ++ E
Sbjct: 399 DVERLLKKQRE 409
>GLCA_SOYBN (P13916) Beta-conglycinin, alpha chain precursor
Length = 605
Score = 400 bits (1028), Expect = e-111
Identities = 198/366 (54%), Positives = 283/366 (77%), Gaps = 18/366 (4%)
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF+F SNRF+TLF+N+ G IR+LQRF++RS +NL++YR+LE++SKP+TL LP H D
Sbjct: 193 KNPFLFGSNRFETLFKNQYGRIRVLQRFNQRSPQLQNLRDYRILEFNSKPNTLLLPNHAD 252
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIP 193
AD+++V+++G AIL+++N ++R+S+ L+ GD +++P+GT Y+ N D+N++LR++ LAIP
Sbjct: 253 ADYLIVILNGTAILSLVNNDDRDSYRLQSGDALRVPSGTTYYVVNPDNNENLRLITLAIP 312
Query: 194 VNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLR 253
VN+PG+F+SF LS +E QQS+L GFS+NILEA++++ +EEI +VL E G +
Sbjct: 313 VNKPGRFESFFLSSTEAQQSYLQGFSRNILEASYDTKFEEINKVLFSREE-------GQQ 365
Query: 254 KDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNF 313
+ E+R Q +VIV++S+EQI LSK AKSSSR++ SSE +P NLR++ PIYSNK G F
Sbjct: 366 QGEQRLQE---SVIVEISKEQIRALSKRAKSSSRKTISSEDKPFNLRSRDPIYSNKLGKF 422
Query: 314 FEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQ 373
FEITPEKNPQL+DLDI ++ ++ EG+LLLPHFNS+A VI+ + EG ELVG + E Q
Sbjct: 423 FEITPEKNPQLRDLDIFLSIVDMNEGALLLPHFNSKAIVILVINEGDANIELVGLK-EQQ 481
Query: 374 QEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENN 433
QEQ QQ+E+ +V++YRA LS D++VIPAG+P+VV A+S+L+ GINAENN
Sbjct: 482 QEQ-------QQEEQPLEVRKYRAELSEQDIFVIPAGYPVVVNATSNLNFFAIGINAENN 534
Query: 434 QRNFLA 439
QRNFLA
Sbjct: 535 QRNFLA 540
>AL12_ARAHY (P43238) Allergen Ara h 1, clone P41B precursor (Ara h
I)
Length = 626
Score = 397 bits (1021), Expect = e-110
Identities = 199/379 (52%), Positives = 270/379 (70%), Gaps = 7/379 (1%)
Query: 68 RSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLF 127
R + + NPF F S RF T + N+NG IR+LQRFD+RS+ F+NLQN+R+++ +KP+TL
Sbjct: 166 REETSRNNPFYFPSRRFSTRYGNQNGRIRVLQRFDQRSRQFQNLQNHRIVQIEAKPNTLV 225
Query: 128 LPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRV 187
LP+H DAD ILV+ G+A +TV N NNR SFNL+ G +++P+G + Y+ NR DN++LRV
Sbjct: 226 LPKHADADNILVIQQGQATVTVANGNNRKSFNLDEGHALRIPSGFISYILNRHDNQNLRV 285
Query: 188 LDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQ 247
+++PVN PGQF+ F + S +Q S+L GFS+N LEAAFN+ + EI RVL+EEN Q
Sbjct: 286 AKISMPVNTPGQFEDFFPASSRDQSSYLQGFSRNTLEAAFNAEFNEIRRVLLEENAGGEQ 345
Query: 248 HRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESE---PINLRNQKP 304
RG R+ R VIVKVS+E +EEL+K+AKS S++ E + PINLR +P
Sbjct: 346 EERGQRRWSTRSSENNEGVIVKVSKEHVEELTKHAKSVSKKGSEEEGDITNPINLREGEP 405
Query: 305 IYSNKFGNFFEITPE-KNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEF 363
SN FG FE+ P+ KNPQL+DLD+++ EI+EG+L+LPHFNS+A VIV V +G G
Sbjct: 406 DLSNNFGKLFEVKPDKKNPQLQDLDMMLTCVEIKEGALMLPHFNSKAMVIVVVNKGTGNL 465
Query: 364 ELVGQRNENQQE-QREYEEDEQQQER--SQQVQRYRARLSPGDVYVIPAGHPIVVTASSD 420
ELV R E QQ +RE EEDE ++E +++V+RY ARL GDV+++PA HP+ + ASS+
Sbjct: 466 ELVAVRKEQQQRGRREEEEDEDEEEEGSNREVRRYTARLKEGDVFIMPAAHPVAINASSE 525
Query: 421 LSLLGFGINAENNQRNFLA 439
L LLGFGINAENN R FLA
Sbjct: 526 LHLLGFGINAENNHRIFLA 544
>AL11_ARAHY (P43237) Allergen Ara h 1, clone P17 precursor (Ara h I)
Length = 614
Score = 395 bits (1014), Expect = e-109
Identities = 198/381 (51%), Positives = 271/381 (70%), Gaps = 10/381 (2%)
Query: 68 RSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLF 127
R + + NPF F S RF T + N+NG IR+LQRFD+RSK F+NLQN+R+++ ++P+TL
Sbjct: 160 REETSRNNPFYFPSRRFSTRYGNQNGRIRVLQRFDQRSKQFQNLQNHRIVQIEARPNTLV 219
Query: 128 LPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRV 187
LP+H DAD ILV+ G+A +TV N NNR SFNL+ G +++P+G + Y+ NR DN++LRV
Sbjct: 220 LPKHADADNILVIQQGQATVTVANGNNRKSFNLDEGHALRIPSGFISYILNRHDNQNLRV 279
Query: 188 LDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQ 247
+++PVN PGQF+ F + S +Q S+L GFS+N LEAAFN+ + EI RVL+EEN Q
Sbjct: 280 AKISMPVNTPGQFEDFFPASSRDQSSYLQGFSRNTLEAAFNAEFNEIRRVLLEENAGGEQ 339
Query: 248 HRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSE--SEPINLRNQKPI 305
RG R+ R E VIVKVS+E ++EL+K+AKS S++ E + PINLR+ +P
Sbjct: 340 EERGQRRRSTRSSDNE-GVIVKVSKEHVQELTKHAKSVSKKGSEEEDITNPINLRDGEPD 398
Query: 306 YSNKFGNFFEITPE-KNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFE 364
SN FG FE+ P+ KNPQL+DLD+++ EI+EG+L+LPHFNS+A VIV V +G G E
Sbjct: 399 LSNNFGRLFEVKPDKKNPQLQDLDMMLTCVEIKEGALMLPHFNSKAMVIVVVNKGTGNLE 458
Query: 365 LVGQRNENQ------QEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTAS 418
LV R E Q QE E EEDE+++ +++V+RY ARL GDV+++PA HP+ + AS
Sbjct: 459 LVAVRKEQQQRGRREQEWEEEEEDEEEEGSNREVRRYTARLKEGDVFIMPAAHPVAINAS 518
Query: 419 SDLSLLGFGINAENNQRNFLA 439
S+L LLGFGINAENN R FLA
Sbjct: 519 SELHLLGFGINAENNHRIFLA 539
>GLCX_SOYBN (P11827) Beta-conglycinin, alpha' chain precursor
Length = 639
Score = 382 bits (982), Expect = e-106
Identities = 192/385 (49%), Positives = 284/385 (72%), Gaps = 38/385 (9%)
Query: 74 ENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHND 133
+NPF FNS RFQTLF+N+ GH+R+LQRF+KRS+ +NL++YR+LE++SKP+TL LP H D
Sbjct: 209 KNPFHFNSKRFQTLFKNQYGHVRVLQRFNKRSQQLQNLRDYRILEFNSKPNTLLLPHHAD 268
Query: 134 ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLP-------------------AGTLG 174
AD+++V+++G AILT++N ++R+S+NL+ GD +++P AGT
Sbjct: 269 ADYLIVILNGTAILTLVNNDDRDSYNLQSGDALRVPAGTTFYVVNPDNDENLRMIAGTTF 328
Query: 175 YLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEI 234
Y+ N D++++LR++ LAIPVN+PG+F+SF LS ++ QQS+L GFSKNILEA++++ +EEI
Sbjct: 329 YVVNPDNDENLRMITLAIPVNKPGRFESFFLSSTQAQQSYLQGFSKNILEASYDTKFEEI 388
Query: 235 ERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSES 294
+VL E + Q +ER Q+S VIV++S++QI ELSK+AKSSSR++ SSE
Sbjct: 389 NKVLFGREEGQQQ------GEERLQES----VIVEISKKQIRELSKHAKSSSRKTISSED 438
Query: 295 EPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIV 354
+P NL ++ PIYSNK G FEIT ++NPQL+DLD+ ++ ++ EG+L LPHFNS+A V++
Sbjct: 439 KPFNLGSRDPIYSNKLGKLFEIT-QRNPQLRDLDVFLSVVDMNEGALFLPHFNSKAIVVL 497
Query: 355 AVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIV 414
+ EG+ ELVG + + Q+ +QQ+E+ +V++YRA LS D++VIPAG+P++
Sbjct: 498 VINEGEANIELVGIKEQQQR--------QQQEEQPLEVRKYRAELSEQDIFVIPAGYPVM 549
Query: 415 VTASSDLSLLGFGINAENNQRNFLA 439
V A+SDL+ FGINAENNQRNFLA
Sbjct: 550 VNATSDLNFFAFGINAENNQRNFLA 574
Score = 35.0 bits (79), Expect = 0.38
Identities = 37/153 (24%), Positives = 67/153 (43%), Gaps = 25/153 (16%)
Query: 126 LFLPQHND-ADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
LFLP N A +LV+ G+A + ++ + +R + P Y A + +D
Sbjct: 484 LFLPHFNSKAIVVLVINEGEANIELVGIKEQQ----QRQQQEEQPLEVRKYRAELSE-QD 538
Query: 185 LRVLDLAIPV----NRPGQFQSFSLSESENQQSFLSGFSKNIL--------EAAFNSNYE 232
+ V+ PV F +F ++ NQ++FL+G N++ E AF + +
Sbjct: 539 IFVIPAGYPVMVNATSDLNFFAFGINAENNQRNFLAGSKDNVISQIPSQVQELAFPRSAK 598
Query: 233 EIERVLIEENEQEPQHRRGLRKDERRQQSQEAN 265
+IE ++ ++E D + QQ +E N
Sbjct: 599 DIENLIKSQSE-------SYFVDAQPQQKEEGN 624
>CANA_CANGL (P10562) Canavalin precursor
Length = 445
Score = 372 bits (956), Expect = e-103
Identities = 190/411 (46%), Positives = 281/411 (68%), Gaps = 45/411 (10%)
Query: 43 MAIKAPCSLLMLLGIVFLASICVS--------------SRSDQDQENPFIFNSNRFQTLF 88
MA A L +LLG+V LAS+ S S + Q NP++F SN+F TLF
Sbjct: 1 MAFSARFPLWLLLGVVLLASVSASFAHSGHSGGEAEDESEESRAQNNPYLFRSNKFLTLF 60
Query: 89 ENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILT 148
+N++G +RLLQRF++ ++ ENL++YR+LEY SKP+TL LP H+D+D +++V+ G+AIL
Sbjct: 61 KNQHGSLRLLQRFNEDTEKLENLRDYRVLEYCSKPNTLLLPHHSDSDLLVLVLEGQAILV 120
Query: 149 VLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSES 208
++NP+ R+++ L++GD IK+ AGT YL N D+N++LR+L+ AI RPG + F LS +
Sbjct: 121 LVNPDGRDTYKLDQGDAIKIQAGTPFYLINPDNNQNLRILNFAITFRRPGTVEDFFLSST 180
Query: 209 ENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIV 268
+ S+LS FSKN LEA+++S Y+EIE+ L++E ++ VIV
Sbjct: 181 KRLPSYLSAFSKNFLEASYDSPYDEIEQTLLQEEQE--------------------GVIV 220
Query: 269 KVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLD 328
K+ ++QI+E+SK+A+SSSR++ SS+ +P NLR++ PIYSN +G +EITPEKN QL+DLD
Sbjct: 221 KMPKDQIQEISKHAQSSSRKTLSSQDKPFNLRSRDPIYSNNYGKLYEITPEKNSQLRDLD 280
Query: 329 ILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQER 388
IL+N ++ EG+L +PH+NSRATVI+ EG+ E ELVG E+ +QQ
Sbjct: 281 ILLNCLQMNEGALFVPHYNSRATVILVANEGRAEVELVG-----------LEQQQQQGLE 329
Query: 389 SQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
S Q++RY A LS GD+ VIP+ P+ + A+SDL+++G G+NAENN+RNFLA
Sbjct: 330 SMQLRRYAATLSEGDILVIPSSFPVALKAASDLNMVGIGVNAENNERNFLA 380
>CANA_CANEN (P50477) Canavalin precursor
Length = 445
Score = 372 bits (954), Expect = e-102
Identities = 190/411 (46%), Positives = 280/411 (67%), Gaps = 45/411 (10%)
Query: 43 MAIKAPCSLLMLLGIVFLASICVS--------------SRSDQDQENPFIFNSNRFQTLF 88
MA A L +LLG+V LAS+ S S + Q NP++F SN+F TLF
Sbjct: 1 MAFSARFPLWLLLGVVLLASVSASFAHSGHSGGEAEDESEESRAQNNPYLFRSNKFLTLF 60
Query: 89 ENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILT 148
+N++G +RLLQRF++ ++ ENL++YR+LEY SKP+TL LP H+D+D +++V+ G+AIL
Sbjct: 61 KNQHGSLRLLQRFNEDTEKLENLRDYRVLEYCSKPNTLLLPHHSDSDLLVLVLEGQAILV 120
Query: 149 VLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSES 208
++NP+ R+++ L++GD IK+ AGT YL N D+N++LR+L AI RPG + F LS +
Sbjct: 121 LVNPDGRDTYKLDQGDAIKIQAGTPFYLINPDNNQNLRILKFAITFRRPGTVEDFFLSST 180
Query: 209 ENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIV 268
+ S+LS FSKN LEA+++S Y+EIE+ L++E ++ VIV
Sbjct: 181 KRLPSYLSAFSKNFLEASYDSPYDEIEQTLLQEEQE--------------------GVIV 220
Query: 269 KVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLD 328
K+ ++QI+E+SK+A+SSSR++ SS+ +P NLR++ PIYSN +G +EITPEKN QL+DLD
Sbjct: 221 KMPKDQIQEISKHAQSSSRKTLSSQDKPFNLRSRDPIYSNNYGKLYEITPEKNSQLRDLD 280
Query: 329 ILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQER 388
IL+N ++ EG+L +PH+NSRATVI+ EG+ E ELVG E+ +QQ
Sbjct: 281 ILLNCLQMNEGALFVPHYNSRATVILVANEGRAEVELVG-----------LEQQQQQGLE 329
Query: 389 SQQVQRYRARLSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
S Q++RY A LS GD+ VIP+ P+ + A+SDL+++G G+NAENN+RNFLA
Sbjct: 330 SMQLRRYAATLSEGDIIVIPSSFPVALKAASDLNMVGIGVNAENNERNFLA 380
>PHSA_PHAVU (P07219) Phaseolin, alpha-type precursor
Length = 436
Score = 313 bits (801), Expect = 7e-85
Identities = 184/401 (45%), Positives = 251/401 (61%), Gaps = 57/401 (14%)
Query: 52 LMLLGIVFLASICVS-----SRSDQDQENPFIFNS-NRFQTLFENENGHIRLLQRFDKRS 105
L+LLGI+FLAS+ S ++ Q+NPF FNS N + TLF+N+ GHIR+LQRFD++S
Sbjct: 8 LLLLGILFLASLSASFATSLREEEESQDNPFYFNSDNSWNTLFKNQYGHIRVLQRFDQQS 67
Query: 106 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNS-FNLERG- 163
K +NL++YRL+E+ SKP TL LPQ DA+ +LVV SG AIL ++ P++R F L +G
Sbjct: 68 KRLQNLEDYRLVEFRSKPETLLLPQQADAELLLVVRSGSAILVLVKPDDRREYFFLTQGD 127
Query: 164 -----DTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGF 218
D K+PAGT+ YL N D +DLR++ LA+PVN P Q F LS +E QQS+L F
Sbjct: 128 NPIFSDNQKIPAGTIFYLVNPDPKEDLRIIQLAMPVNNP-QIHEFFLSSTEAQQSYLQEF 186
Query: 219 SKNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEEL 278
SK+ILEA+FNS +EEI RVL EE Q+ ++ Q+ VIV + EQIEEL
Sbjct: 187 SKHILEASFNSKFEEINRVLFEEEGQQ-------------EEGQQEGVIVNIDSEQIEEL 233
Query: 279 SKNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIRE 338
SK+AKSSSR+S S + I N+FGN E T L++L++ E++E
Sbjct: 234 SKHAKSSSRKSHSKQDNTI---------GNEFGNLTERTD------NSLNVLISSIEMKE 278
Query: 339 GSLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRAR 398
G+L +PH+ S+A VI+ V EG+ ELVG + +E E+E YRA
Sbjct: 279 GALFVPHYYSKAIVILVVNEGEAHVELVGPK--GNKETLEFES-------------YRAE 323
Query: 399 LSPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
LS DV+VIPA +P+ + A+S+++ GFGINA NN RN LA
Sbjct: 324 LSKDDVFVIPAAYPVAIKATSNVNFTGFGINANNNNRNLLA 364
>PHSB_PHAVU (P02853) Phaseolin, beta-type precursor
Length = 421
Score = 305 bits (781), Expect = 2e-82
Identities = 181/400 (45%), Positives = 245/400 (61%), Gaps = 61/400 (15%)
Query: 52 LMLLGIVFLASICVS-----SRSDQDQENPFIFNS-NRFQTLFENENGHIRLLQRFDKRS 105
L+LLGI+FLAS+ S ++ Q+NPF FNS N + TLF+N+ GHIR+LQRFD++S
Sbjct: 8 LLLLGILFLASLSASFATSLREEEESQDNPFYFNSDNSWNTLFKNQYGHIRVLQRFDQQS 67
Query: 106 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSF------N 159
K +NL++YRL+E+ SKP TL LPQ DA+ +LVV SG AIL ++ P++R + N
Sbjct: 68 KRLQNLEDYRLVEFRSKPETLLLPQQADAELLLVVRSGSAILVLVKPDDRREYFFLTSDN 127
Query: 160 LERGDTIKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFS 219
D K+PAGT+ YL N D +DLR++ LA+PVN P Q F LS +E QQS+L FS
Sbjct: 128 PIFSDHQKIPAGTIFYLVNPDPKEDLRIIQLAMPVNNP-QIHEFFLSSTEAQQSYLQEFS 186
Query: 220 KNILEAAFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELS 279
K+ILEA+FNS +EEI RVL EE + Q+ VIV + EQI+ELS
Sbjct: 187 KHILEASFNSKFEEINRVLFEE------------------EGQQEGVIVNIDSEQIKELS 228
Query: 280 KNAKSSSRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREG 339
K+AKSSSR+S S + I N+FGN E T L++L++ E+ EG
Sbjct: 229 KHAKSSSRKSLSKQDNTI---------GNEFGNLTERTD------NSLNVLISSIEMEEG 273
Query: 340 SLLLPHFNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARL 399
+L +PH+ S+A VI+ V EG+ ELVG + +E EYE YRA L
Sbjct: 274 ALFVPHYYSKAIVILVVNEGEAHVELVGPK--GNKETLEYES-------------YRAEL 318
Query: 400 SPGDVYVIPAGHPIVVTASSDLSLLGFGINAENNQRNFLA 439
S DV+VIPA +P+ + A+S+++ GFGINA NN RN LA
Sbjct: 319 SKDDVFVIPAAYPVAIKATSNVNFTGFGINANNNNRNLLA 358
>PHS2_PHALU (Q43617) Phaseolin precursor (Fragment)
Length = 423
Score = 301 bits (770), Expect = 3e-81
Identities = 173/393 (44%), Positives = 248/393 (63%), Gaps = 55/393 (13%)
Query: 52 LMLLGIVFLASICVS-----SRSDQDQENPFIFNS-NRFQTLFENENGHIRLLQRFDKRS 105
L+LLGI+FLAS+ S ++ Q+NPF F+S N +QTLF+N+ GHIR+LQRFD+RS
Sbjct: 5 LLLLGILFLASLSASFAISLREHNESQDNPFYFSSDNSWQTLFKNQYGHIRVLQRFDQRS 64
Query: 106 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDT 165
+ +NL++YRL+E+ SKP TL LPQ DA+F+LVV SG A+L ++ P ++L++ DT
Sbjct: 65 ERLQNLEDYRLVEFMSKPETLLLPQQADAEFLLVVRSGSALLALVKPGGTIIYSLKQQDT 124
Query: 166 IKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEA 225
+K+PAGT+ +L N ++N+DLR++ LA+ VN P Q Q F LS +E QQS+L GFSK+IL+A
Sbjct: 125 LKIPAGTIFFLINPENNEDLRIIKLAMTVNNP-QIQDFFLSSTEAQQSYLYGFSKHILDA 183
Query: 226 AFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSS 285
+FNS E+I R+L E + ++ VIV + + I+ELS++AKSS
Sbjct: 184 SFNSPIEKINRLLFAE------------------EGRQEGVIVNIGSDLIQELSRHAKSS 225
Query: 286 SRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPH 345
SR+S S I SN++GN +I LD+L+ Y EI+EG L +PH
Sbjct: 226 SRKSLDHNSLDI---------SNEWGNLTDIV------YNSLDVLLTYVEIKEGGLFVPH 270
Query: 346 FNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVY 405
+NS+A VI+ VEEG + ELVG + E + S +++ YRA +S GDV+
Sbjct: 271 YNSKAIVILVVEEGVAKVELVGPKREKE---------------SLELETYRADVSEGDVF 315
Query: 406 VIPAGHPIVVTASSDLSLLGFGINAENNQRNFL 438
VIPA P + A S+++ FGINA NN R FL
Sbjct: 316 VIPAAFPFAIKAISNVNFTSFGINANNNYRIFL 348
>VCLA_PEA (P02855) Provicilin (Type A) (Fragment)
Length = 275
Score = 298 bits (764), Expect = 1e-80
Identities = 148/210 (70%), Positives = 182/210 (86%), Gaps = 5/210 (2%)
Query: 233 EIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESS 292
EIE++L+EE+E+E HRRGLR ++RQQSQE NVIVKVS++QIEELSKNAKSSS++S SS
Sbjct: 4 EIEKILLEEHEKETHHRRGLR--DKRQQSQEKNVIVKVSKKQIEELSKNAKSSSKKSVSS 61
Query: 293 ESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATV 352
SEP NL++ PIYSN++G FFEITP+KNPQL+DLDI VNY EI+EGSL LPH+NSRA V
Sbjct: 62 RSEPFNLKSSDPIYSNQYGKFFEITPKKNPQLQDLDIFVNYVEIKEGSLWLPHYNSRAIV 121
Query: 353 IVAVEEGKGEFELVGQRNENQQ---EQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPA 409
IV V EGKG+FELVGQRNENQQ E+ + EE+++++E QVQ Y+A+L+PGDV+VIPA
Sbjct: 122 IVTVNEGKGDFELVGQRNENQQGLREEDDEEEEQREEETKNQVQSYKAKLTPGDVFVIPA 181
Query: 410 GHPIVVTASSDLSLLGFGINAENNQRNFLA 439
GHP+ V ASS+L+LLGFGINAENNQRNFLA
Sbjct: 182 GHPVAVRASSNLNLLGFGINAENNQRNFLA 211
>PHS1_PHALU (P80463) Phaseolin precursor
Length = 428
Score = 295 bits (756), Expect = 1e-79
Identities = 170/393 (43%), Positives = 245/393 (62%), Gaps = 55/393 (13%)
Query: 52 LMLLGIVFLASICVS-----SRSDQDQENPFIFNS-NRFQTLFENENGHIRLLQRFDKRS 105
L+LLGI+FLAS+ S ++ Q+NPF F+S N +QTLF+N+ GHIR+LQ FD+ S
Sbjct: 8 LLLLGILFLASLSASFAISLREHNESQDNPFYFSSDNSWQTLFKNQYGHIRVLQSFDQHS 67
Query: 106 KIFENLQNYRLLEYHSKPHTLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDT 165
+ +NL++YRL+E+ SKP TL LPQ DA+F+LVV SG A+L ++ P ++L++ DT
Sbjct: 68 ERLQNLEDYRLVEFMSKPETLLLPQQADAEFLLVVRSGSALLALVKPGGTIIYSLKQQDT 127
Query: 166 IKLPAGTLGYLANRDDNKDLRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEA 225
+K+PAGT+ +L N +N+DLR++ LA+ VN P Q Q F LS +E QQS+L GF K+IL+A
Sbjct: 128 LKIPAGTIFFLINPQNNEDLRIIKLAMTVNNP-QIQDFFLSSTEAQQSYLYGFRKDILDA 186
Query: 226 AFNSNYEEIERVLIEENEQEPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSS 285
+FNS EEI R+L E + ++ VIV + + I+ELS++AKSS
Sbjct: 187 SFNSPIEEINRLLFAE------------------EGRQEGVIVNIGSDLIQELSRHAKSS 228
Query: 286 SRRSESSESEPINLRNQKPIYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPH 345
SR+S S I SN++GN +I LD+L+ Y EI+EG L +PH
Sbjct: 229 SRKSLDHNSLDI---------SNEWGNLTDIV------YNSLDVLLTYVEIKEGGLFVPH 273
Query: 346 FNSRATVIVAVEEGKGEFELVGQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVY 405
+NS+A VI+ VEEG + ELVG + E + S +++ YRA +S GDV+
Sbjct: 274 YNSKAIVILVVEEGVAKVELVGPKREKE---------------SLELETYRADVSEGDVF 318
Query: 406 VIPAGHPIVVTASSDLSLLGFGINAENNQRNFL 438
VIPA +P+ + A S+++ FGINA NN R L
Sbjct: 319 VIPAAYPVAIKAISNVNFTSFGINANNNYRILL 351
>CVCB_PEA (P13919) Convicilin precursor (Fragment)
Length = 386
Score = 275 bits (703), Expect = 2e-73
Identities = 135/205 (65%), Positives = 169/205 (81%), Gaps = 11/205 (5%)
Query: 67 SRSDQDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTL 126
S Q++ NPF+F SN+F TLFENENGHIRLLQRFDKRS +FENLQNYRL+EY +KPHT+
Sbjct: 193 SSESQERRNPFLFKSNKFLTLFENENGHIRLLQRFDKRSDLFENLQNYRLVEYRAKPHTI 252
Query: 127 FLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLR 186
FLPQH DAD ILVV+SGKAILTVL+PN+RNS+NLERGDTIKLPAGT YL N+DD +DLR
Sbjct: 253 FLPQHIDADLILVVLSGKAILTVLSPNDRNSYNLERGDTIKLPAGTTSYLVNQDDEEDLR 312
Query: 187 VLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEP 246
++DL IPVN PG+F++F L++++NQ +L GFSKNILEA++N+ YE IE+VL+EE E++
Sbjct: 313 LVDLVIPVNGPGKFEAFDLAKNKNQ--YLRGFSKNILEASYNTRYETIEKVLLEEQEKD- 369
Query: 247 QHRRGLRKDERRQQSQEANVIVKVS 271
+RRQQ +E + IVKVS
Sbjct: 370 --------RKRRQQGEETDAIVKVS 386
>VCLA_GOSHI (P09799) Vicilin GC72-A precursor (Alpha-globulin A)
Length = 605
Score = 259 bits (663), Expect = 7e-69
Identities = 151/378 (39%), Positives = 222/378 (57%), Gaps = 25/378 (6%)
Query: 71 QDQENPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLPQ 130
Q + NP+ F+ FQ F E+G+ R+LQRF + + + +R+ + P+T LP
Sbjct: 177 QQRNNPYYFHRRSFQERFREEHGNFRVLQRFADKHHLLRGINEFRIAILEANPNTFVLPH 236
Query: 131 HNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLDL 190
H DA+ I VV +G+ +T + N+ S+N+ G +++PAG+ YLAN+D+ + L + L
Sbjct: 237 HCDAEKIYVVTNGRGTVTFVTHENKESYNVVPGVVVRIPAGSTVYLANQDNREKLTIAVL 296
Query: 191 AIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHRR 250
PVN PGQFQ F + EN QS+L FS+ ILEA FN+ E+++ +
Sbjct: 297 HRPVNNPGQFQKFFPAGQENPQSYLRIFSREILEAVFNTRSEQLDEL------------P 344
Query: 251 GLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNKF 310
G R+ RRQQ Q + K S+EQI LS+ A +S R + SE NL +Q P YSN+
Sbjct: 345 GGRQSHRRQQGQ--GMFRKASQEQIRALSQGA--TSPRGKGSEGYAFNLLSQTPRYSNQN 400
Query: 311 GNFFEITPEK-NPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELVG-- 367
G F+E P QL+++D V EI +GS+ +PH+NS+AT +V V EG G E+V
Sbjct: 401 GRFYEACPRNFQQQLREVDSSVVAFEINKGSIFVPHYNSKATFVVLVTEGNGHVEMVCPH 460
Query: 368 -QRNENQQEQREYEEDEQQ--QERSQQVQRYRARLSPGDVYVIPAGHPIVVTAS--SDLS 422
R + RE EE E+Q + RS Q +R RA+LS G+++V+PAGHP+ AS DL
Sbjct: 461 LSRQSSDWSSREEEEQEEQEVERRSGQYKRVRAQLSTGNLFVVPAGHPVTFVASQNEDLG 520
Query: 423 LLGFGI-NAENNQRNFLA 439
LLGFG+ N ++N+R F+A
Sbjct: 521 LLGFGLYNGQDNKRIFVA 538
>VCL_THECC (Q43358) Vicilin precursor
Length = 525
Score = 256 bits (653), Expect = 1e-67
Identities = 135/379 (35%), Positives = 225/379 (58%), Gaps = 25/379 (6%)
Query: 71 QDQENPFIFNSNR-FQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPHTLFLP 129
Q + NP+ F R FQT F +E G+ ++LQRF + S + + +YRL + + P+T LP
Sbjct: 138 QQRNNPYYFPKRRSFQTRFRDEEGNFKILQRFAENSPPLKGINDYRLAMFEANPNTFILP 197
Query: 130 QHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKDLRVLD 189
H DA+ I V +GK +T + N+ S+N++RG + +PAG+ Y+ ++D+ + L +
Sbjct: 198 HHCDAEAIYFVTNGKGTITFVTHENKESYNVQRGTVVSVPAGSTVYVVSQDNQEKLTIAV 257
Query: 190 LAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQEPQHR 249
LA+PVN PG+++ F + + +S+ FS +LE FN+ E++E +L E+
Sbjct: 258 LALPVNSPGKYELFFPAGNNKPESYYGAFSYEVLETVFNTQREKLEEILEEQ-------- 309
Query: 250 RGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKPIYSNK 309
+ ++RQQ Q+ + K EQI +S+ A +S R E INL +Q P+YSN+
Sbjct: 310 ----RGQKRQQGQQ-GMFRKAKPEQIRAISQQA--TSPRHRGGERLAINLLSQSPVYSNQ 362
Query: 310 FGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFELV--- 366
G FFE PE Q +++D+ V+ ++ +G++ +PH+NS+AT +V V +G G ++
Sbjct: 363 NGRFFEACPEDFSQFQNMDVAVSAFKLNQGAIFVPHYNSKATFVVFVTDGYGYAQMACPH 422
Query: 367 ----GQRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTASSD-- 420
Q +++ ++ R +E+E ++E + Q+ +A LSPGDV+V PAGH + AS D
Sbjct: 423 LSRQSQGSQSGRQDRREQEEESEEETFGEFQQVKAPLSPGDVFVAPAGHAVTFFASKDQP 482
Query: 421 LSLLGFGINAENNQRNFLA 439
L+ + FG+NA+NNQR FLA
Sbjct: 483 LNAVAFGLNAQNNQRIFLA 501
>VCLB_GOSHI (P09801) Vicilin C72 precursor (Alpha-globulin B)
Length = 588
Score = 253 bits (646), Expect = 7e-67
Identities = 143/387 (36%), Positives = 228/387 (57%), Gaps = 29/387 (7%)
Query: 66 SSRSDQDQE-NPFIFNSNRFQTLFENENGHIRLLQRFDKRSKIFENLQNYRLLEYHSKPH 124
+ +Q+Q NPF F+ FQ+ F E+G+ R+LQRF R I + +RL + P+
Sbjct: 170 TEEGEQEQSHNPFHFHRRSFQSRFREEHGNFRVLQRFASRHPILRGINEFRLSILEANPN 229
Query: 125 TLFLPQHNDADFILVVVSGKAILTVLNPNNRNSFNLERGDTIKLPAGTLGYLANRDDNKD 184
T LP H DA+ I +V +G+ LT L N+ S+N+ G +++PAG+ YLAN+D+ +
Sbjct: 230 TFVLPHHCDAEKIYLVTNGRGTLTFLTHENKESYNVVPGVVVRVPAGSTVYLANQDNKEK 289
Query: 185 LRVLDLAIPVNRPGQFQSFSLSESENQQSFLSGFSKNILEAAFNSNYEEIERVLIEENEQ 244
L + L PVN P QF+ F + S+ QS+L FS+ ILE AFN+ E+++ +
Sbjct: 290 LIIAVLHRPVNNPRQFEEFFPAGSQRPQSYLRAFSREILEPAFNTRSEQLDELF------ 343
Query: 245 EPQHRRGLRKDERRQQSQEANVIVKVSREQIEELSKNAKSSSRRSESSESEPINLRNQKP 304
G R+ RRQQ Q + K S+EQI LS+ A +S R +S E NL + P
Sbjct: 344 ------GGRQSHRRQQGQ--GMFRKASQEQIRALSQEA--TSPREKSGERFAFNLLYRTP 393
Query: 305 IYSNKFGNFFEITPEKNPQLKDLDILVNYAEIREGSLLLPHFNSRATVIVAVEEGKGEFE 364
YSN+ G F+E P + QL D+++ V+ ++ +GS+ +PH+NS+AT +V V EG G E
Sbjct: 394 RYSNQNGRFYEACPREFRQLSDINVTVSALQLNQGSIFVPHYNSKATFVVLVNEGNGYVE 453
Query: 365 LVG-----QRNENQQEQREYEEDEQQQERSQQVQRYRARLSPGDVYVIPAGHPIVVTAS- 418
+V Q + ++E+++ E++++++ RS Q ++ R++LS GD++V+PA P+ AS
Sbjct: 454 MVSPHLPRQSSFEEEEEQQQEQEQEEERRSGQYRKIRSQLSRGDIFVVPANFPVTFVASQ 513
Query: 419 -SDLSLLGFG-----INAENNQRNFLA 439
+L + GFG IN ++NQR F+A
Sbjct: 514 NQNLRMTGFGLYNQNINPDHNQRIFVA 540
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.133 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,769,006
Number of Sequences: 164201
Number of extensions: 2092181
Number of successful extensions: 15075
Number of sequences better than 10.0: 359
Number of HSP's better than 10.0 without gapping: 114
Number of HSP's successfully gapped in prelim test: 248
Number of HSP's that attempted gapping in prelim test: 12245
Number of HSP's gapped (non-prelim): 1543
length of query: 440
length of database: 59,974,054
effective HSP length: 113
effective length of query: 327
effective length of database: 41,419,341
effective search space: 13544124507
effective search space used: 13544124507
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC148289.2


