

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.8 + phase: 0
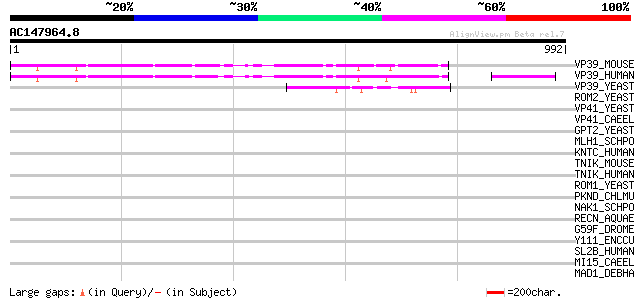
(992 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VP39_MOUSE (Q8R5L3) Vam6/Vps39-like protein (Vps39 protein) 279 3e-74
VP39_HUMAN (Q96JC1) Vam6/Vps39-like protein (hVam6p) 278 7e-74
VP39_YEAST (Q07468) Vacuolar assembly protein VPS39 (Vacuolar mo... 70 4e-11
ROM2_YEAST (P51862) RHO1 GDP-GTP exchange protein 2 43 0.004
VP41_YEAST (P38959) Vacuolar assembly protein VPS41 (Vacuolar mo... 39 0.053
VP41_CAEEL (Q19954) Vacuolar assembly protein VPS41 homolog 39 0.090
GPT2_YEAST (P36148) Glycerol-3-phosphate O-acyltransferase 2 (EC... 37 0.34
MLH1_SCHPO (Q9P7W6) Putative MUTL protein homolog 1 (DNA mismatc... 36 0.45
KNTC_HUMAN (P50748) Kinetochore-associated protein 1 (Rough deal... 36 0.58
TNIK_MOUSE (P83510) Traf2 and NCK interacting kinase (EC 2.7.1.3... 35 0.99
TNIK_HUMAN (Q9UKE5) TRAF2 and NCK interacting kinase (EC 2.7.1.37) 35 0.99
ROM1_YEAST (P53046) RHO1 GDP-GTP exchange protein 1 (Protein kin... 35 0.99
PKND_CHLMU (Q9PK92) Serine/threonine-protein kinase pknD (EC 2.7... 35 1.3
NAK1_SCHPO (O75011) Serine/threonine-protein kinase nak1 (EC 2.7... 34 1.7
RECN_AQUAE (O66834) DNA repair protein recN (Recombination prote... 34 2.2
G59F_DROME (Q9W1N5) Putative gustatory receptor 59f 33 2.9
Y111_ENCCU (Q8STB5) Hypothetical protein ECU01_0110/ECU01_1500/E... 33 3.8
SL2B_HUMAN (Q8NEV8) Slp homolog lacking C2 domains-b (Exophilin 5) 33 3.8
MI15_CAEEL (Q23356) Serine/threonine-protein kinase mig-15 (EC 2... 33 3.8
MAD1_DEBHA (Q6BSY1) Spindle assembly checkpoint component MAD1 (... 33 3.8
>VP39_MOUSE (Q8R5L3) Vam6/Vps39-like protein (Vps39 protein)
Length = 886
Score = 279 bits (713), Expect = 3e-74
Identities = 236/822 (28%), Positives = 392/822 (46%), Gaps = 113/822 (13%)
Query: 2 VHTAYDSFELLTNSTSKIESIESYGSKLLLGCSNGSLLVYAPEHSV-------PAPEEMR 54
+H A++ +L +I+ + ++ LL+G G LL+Y V P
Sbjct: 1 MHDAFEPVPILEKLPLQIDCLAAWEEWLLVGTKQGHLLLYRIRKDVVPADVASPESGSCN 60
Query: 55 KEAYVLERNVNGFAKKAVVSLQVVESREFLLSLSES-IAFHKLPTFETIAVITKAKGANA 113
+ LE++ F+KK + + VV + L+SL E+ I H L TF+ I ++KAKGA+
Sbjct: 61 RFEVTLEKSNKNFSKK-IQQIHVVSQFKILVSLLENNIYVHDLLTFQQITTVSKAKGASL 119
Query: 114 FCWD-------EHRGFLCFARQKRVCI-FRRDGGRGFVEVK-DFGVLDVVKSMSWCGENI 164
F D E +C A +K++ + F +D R F E++ DF V DV KSM+WC +I
Sbjct: 120 FTCDLQHTETGEEVLRMCVAVRKKLQLYFWKD--REFHELQGDFSVPDVPKSMAWCENSI 177
Query: 165 CLGIRKAYVILNATS-GSISEVFTSGRLAPPLVVSLPSGELLLGKDNIGVIVDQNGKLRP 223
C+G ++ Y ++ GSI E+F +G+ PLV L G++ +G+D++ V++++ G
Sbjct: 178 CVGFKRDYYLIRVDGKGSIKELFPTGKQLEPLVAPLADGKVAVGQDDLTVVLNEEGICTQ 237
Query: 224 EGRICWSEAPTEVVIQNPYALALLPRFVEIRSLRGPYPLIQTIVFRNVRHLRQSNNSVI- 282
+ + W++ P + Q PY +A+LPR+VEIR+L P L+Q+I + R + +++I
Sbjct: 238 KCALNWTDIPVAMEHQPPYIVAVLPRYVEIRTLE-PRLLVQSIELQRPRFITSGGSNIIY 296
Query: 283 IALENSIHCLFPVPLGAQIVQLTAAGNFEEALSLCKLLPPEDSNLRAAKEDSIH---IRY 339
+A + + L PVP+ QI QL FE AL L ++ DS K+ IH Y
Sbjct: 297 VASNHFVWRLIPVPMATQIQQLLQDKQFELALQLAEMKDDSDSE----KQQQIHHIKNLY 352
Query: 340 AHYLFDNGSYEESMEHFLASQVDITYVLSLYTSIILPKTTIVHDSDKLDIFGDPLHLSRG 399
A LF ++ESM+ F D T+V+ LY +LP D K + +PL G
Sbjct: 353 AFNLFCQKRFDESMQVFAKLGTDPTHVMGLYPD-LLPT-----DYRKQLQYPNPLPTLSG 406
Query: 400 SSMSDDMEPSSASNMSELDDNAELESKKMSHNMLMALIKFLHKKRHSIIEKATAEGTEEV 459
AELE +ALI +L +KR +++K
Sbjct: 407 ---------------------AELEKAH------LALIDYLTQKRSQLVKKLN------- 432
Query: 460 VFDAVGNNFESYNSNRFKKINKRHGSIPVSSEAREMASILDTALLQAMLLTGQPSMAENL 519
+S+ + P +++ I+DT LL+ L T +A L
Sbjct: 433 ------------DSDHQSSTSPLMEGTPTIKSKKKLLQIIDTTLLKCYLHTNVALVAPLL 480
Query: 520 LRVLNYCDLKICEEILQEGSYHVSLVELYKCNSMHREALEIINKSVKESESSQSKIAHRF 579
N+C ++ E +L++ + L+ LY+ +H +AL+++ V +S+ + S +
Sbjct: 481 RLENNHCHIEESEHVLKKAHKYSELIILYEKKGLHEKALQVL---VDQSKKANSPLKGH- 536
Query: 580 KPEAIIEYLKPLCELDTTLVLEYSMLVLESCPTQTIELFLSG-----NIPADMVNLYLKQ 634
E ++YL+ L + L+ YS+ VL P +++F ++P D V +L +
Sbjct: 537 --ERTVQYLQHLGTENLHLIFSYSVWVLRDFPEDGLKIFTEDLPEVESLPRDRVLNFLIE 594
Query: 635 HAPNLQATYLELVLSMNEGAVSGTLQNEMVHLYLSEVLDWHADL------------SSEQ 682
+ L YLE ++ + E N ++ LY +V D + E+
Sbjct: 595 NFKALAIPYLEHIIHVWE-ETGSQFHNCLIQLYCEKVQSLMKDYLLSLPTGKSPVPAGEE 653
Query: 683 KWDEKVYSPKRKKLLSALESISGYNPEALLKLLPSDALYEERAILLGKMNQHELALSLYV 742
+ Y R+KLL LE S Y+P L+ P D L EERA+LLG+M +HE AL +YV
Sbjct: 654 GGELGEY---RQKLLMFLEISSHYDPGRLICDFPFDGLLEERALLLGRMGKHEQALFIYV 710
Query: 743 HKLHVPELALSYCDHVYESAHKSSVKSLSNIYLMLLQIYLNP 784
H L ++A YC Y+ + + ++YL LL++YL+P
Sbjct: 711 HVLKDTKMAKEYCHKHYDQNKEGN----KDVYLSLLRMYLSP 748
>VP39_HUMAN (Q96JC1) Vam6/Vps39-like protein (hVam6p)
Length = 886
Score = 278 bits (710), Expect = 7e-74
Identities = 238/819 (29%), Positives = 394/819 (48%), Gaps = 107/819 (13%)
Query: 2 VHTAYDSFELLTNSTSKIESIESYGSKLLLGCSNGSLLVYAPEHSV-------PAPEEMR 54
+H A++ +L +I+ + ++ LL+G G LL+Y V P
Sbjct: 1 MHDAFEPVPILEKLPLQIDCLAAWEEWLLVGTKQGHLLLYRIRKDVVPADVASPESGSCN 60
Query: 55 KEAYVLERNVNGFAKKAVVSLQVVESREFLLSLSES-IAFHKLPTFETIAVITKAKGANA 113
+ LE++ F+KK + + VV + L+SL E+ I H L TF+ I ++KAKGA+
Sbjct: 61 RFEVTLEKSNKNFSKK-IQQIHVVSQFKILVSLLENNIYVHDLLTFQQITTVSKAKGASL 119
Query: 114 FCWD-------EHRGFLCFARQKRVCI-FRRDGGRGFVEVK-DFGVLDVVKSMSWCGENI 164
F D E +C A +K++ + F +D R F E++ DF V DV KSM+WC +I
Sbjct: 120 FTCDLQHTETGEEVLQMCVAVKKKLQLYFWKD--REFHELQGDFSVPDVPKSMAWCENSI 177
Query: 165 CLGIRKAYVILNATS-GSISEVFTSGRLAPPLVVSLPSGELLLGKDNIGVIVDQNGKLRP 223
C+G ++ Y ++ GSI E+F +G+ PLV L G++ +G+D++ V++++ G
Sbjct: 178 CVGFKRDYYLIRVDGKGSIKELFPTGKQLEPLVAPLADGKVAVGQDDLTVVLNEEGICTQ 237
Query: 224 EGRICWSEAPTEVVIQNPYALALLPRFVEIRSLRGPYPLIQTIVFRNVRHLRQSNNSVI- 282
+ + W++ P + Q PY +A+LPR+VEIR+ P L+Q+I + R + +++I
Sbjct: 238 KCALNWTDIPVAMEHQPPYIIAVLPRYVEIRTFE-PRLLVQSIELQRPRFITSGGSNIIY 296
Query: 283 IALENSIHCLFPVPLGAQIVQLTAAGNFEEALSLCKLLPPEDSNLRAAKEDSIHI---RY 339
+A + + L PVP+ QI QL FE AL L ++ DS K+ IH Y
Sbjct: 297 VASNHFVWRLIPVPMATQIQQLLQDKQFELALQLAEMKDDSDSE----KQQQIHHIKNLY 352
Query: 340 AHYLFDNGSYEESMEHFLASQVDITYVLSLYTSIILPKTTIVHDSDKLDIFGDPLHLSRG 399
A LF ++ESM+ F D T+V+ LY + LP D K + +PL + G
Sbjct: 353 AFNLFCQKRFDESMQVFAKLGTDPTHVMGLYPDL-LPT-----DYRKQLQYPNPLPVLSG 406
Query: 400 SSMSDDMEPSSASNMSELDDNAELESKKMSHNMLMALIKFLHKKRHSIIEKATAEGTEEV 459
AELE + ALI +L +KR +++K
Sbjct: 407 ---------------------AELEKAHL------ALIDYLTQKRSQLVKKLN------- 432
Query: 460 VFDAVGNNFESYNSNRFKKINKRHGSIPVSSEAREMASILDTALLQAMLLTGQPSMAENL 519
+S+ + P +++ I+DT LL+ L T +A L
Sbjct: 433 ------------DSDHQSSTSPLMEGTPTIKSKKKLLQIIDTTLLKCYLHTNVALVAPLL 480
Query: 520 LRVLNYCDLKICEEILQEGSYHVSLVELYKCNSMHREALEIINKSVKESESSQSKIAHRF 579
N+C ++ E +L++ + L+ LY+ +H +AL+++ V +S+ + S +
Sbjct: 481 RLENNHCHIEESEHVLKKAHKYSELIILYEKKGLHEKALQVL---VDQSKKANSPLKGH- 536
Query: 580 KPEAIIEYLKPLCELDTTLVLEYSMLVLESCPTQTIELFLSG-----NIPADMVNLYLKQ 634
E ++YL+ L + L+ YS+ VL P +++F ++P D V +L +
Sbjct: 537 --ERTVQYLQHLGTENLHLIFSYSVWVLRDFPEDGLKIFTEDLPEVESLPRDRVLGFLIE 594
Query: 635 HAPNLQATYLELVLSMNEGAVSGTLQNEMVHLYLSEV--------LDWHADLSSEQKWDE 686
+ L YLE ++ + E S N ++ LY +V L + A + +E
Sbjct: 595 NFKGLAIPYLEHIIHVWEETGS-RFHNCLIQLYCEKVQGLMKEYLLSFPAGKTPVPAGEE 653
Query: 687 K-VYSPKRKKLLSALESISGYNPEALLKLLPSDALYEERAILLGKMNQHELALSLYVHKL 745
+ R+KLL LE S Y+P L+ P D L EERA+LLG+M +HE AL +YVH L
Sbjct: 654 EGELGEYRQKLLMFLEISSYYDPGRLICDFPFDGLLEERALLLGRMGKHEQALFIYVHIL 713
Query: 746 HVPELALSYCDHVYESAHKSSVKSLSNIYLMLLQIYLNP 784
+A YC H + +K K ++YL LL++YL+P
Sbjct: 714 KDTRMAEEYC-HKHYDRNKDGNK---DVYLSLLRMYLSP 748
Score = 68.2 bits (165), Expect = 1e-10
Identities = 35/114 (30%), Positives = 68/114 (58%), Gaps = 1/114 (0%)
Query: 862 LDEALDLLSRRWDRINGAQALKLLPKETKLQNLLPILGPLVRKSSEMYRNCSVVRSLRQS 921
L AL +L +++ +AL LLP T++ ++ L ++ ++++ R V+++L +
Sbjct: 768 LQAALQVLELHHSKLDTTKALNLLPANTQINDIRIFLEKVLEENAQKKRFNQVLKNLLHA 827
Query: 922 ENLQVKDELYNKRKAVIKISDDNMCSLCHKKIGTSVFAVYPNGKTLVHFVCFRD 975
E L+V++E ++ I+++ +C +C KKIG S FA YPNG +VH+ C ++
Sbjct: 828 EFLRVQEERILHQQVKCIITEEKVCMVCKKKIGNSAFARYPNG-VVVHYFCSKE 880
>VP39_YEAST (Q07468) Vacuolar assembly protein VPS39 (Vacuolar
morphogenesis protein VAM6)
Length = 1049
Score = 69.7 bits (169), Expect = 4e-11
Identities = 81/322 (25%), Positives = 131/322 (40%), Gaps = 48/322 (14%)
Query: 495 MASILDTALLQAMLLTGQPSMAENLLRVLNYCDLKICEEILQEGSYHVSLVELYKCNSMH 554
M +++DT L + L P M +RV N+CD + L+ L++ Y H
Sbjct: 599 MLTLIDTVLFKCYLYYNPP-MVGPFIRVENHCDSHVIVTELKIRHMFKDLIDFYYKRGNH 657
Query: 555 REALEIINKSVKESESSQSKIAHRFKPE-----AIIEYLKPLCELDTTLVLEYSMLVLES 609
EAL+ + V E E+ + R K + +I YLK L ++ Y+ +L
Sbjct: 658 EEALKFLTDLVDELENDNTDQKQRQKIDHGVKILVIYYLKKLSNPQLDVIFTYTDWLLNR 717
Query: 610 CPTQTIELFLSGNIPADM----------VNLYLKQHAPNLQATYLELVLSMNEGAVSGTL 659
+I+ LS D V Y+K+ L YLE +S + L
Sbjct: 718 -HNDSIKEILSSIFFYDSQACSSRDHLKVYGYIKKFDKLLAIQYLEFAIS------TFRL 770
Query: 660 QNEMVHLYLSEVLDWHADLSSEQKWDEKVYSPKRKKLLSALESISGYNPEALLKLLP--- 716
+ +H L ++ + D+ S R KL S LE+ S Y P +LKLL
Sbjct: 771 EGNKLHTVLIKLYLENLDIPST-----------RIKLKSLLETTSVYEPRTILKLLNDAI 819
Query: 717 ---SDALYEE--------RAILLGKMNQHELALSLYVHKLHVPELALSYCDHVYESAHKS 765
SD L + L K+ H+ A+ + + ++ + A SYC+ VY+S
Sbjct: 820 ESGSDQLPTNQLNFVKYLKIFPLSKLENHKEAVHILLDEIDDYKAATSYCNDVYQSDSTK 879
Query: 766 SVKSLSNIYLMLLQIYLNPRRT 787
+ L +Y L+ IY + R +
Sbjct: 880 GEELLLYLYSKLVSIYDSNRNS 901
Score = 37.0 bits (84), Expect = 0.26
Identities = 25/112 (22%), Positives = 54/112 (47%), Gaps = 5/112 (4%)
Query: 866 LDLLSRRWDRINGAQALKLLPKETKLQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENLQ 925
L+ L ++N A+ K LP++ L ++ ++ L++K + + ++L Q E +
Sbjct: 905 LNFLQDHGSKLNSAEIYKNLPQDISLYDIGRVVSQLLKKHTSKMDETRLEKALLQVELVA 964
Query: 926 VKDELYNKRKAVIKISDDNMCSLCHKKI---GTSVFAVYP-NGKTLV-HFVC 972
+L + + +SD + C +C K I GT + + G+ ++ H+ C
Sbjct: 965 TTYKLNERMSSYGVLSDSHKCPICKKVISNFGTDSISWFTREGRNIITHYNC 1016
>ROM2_YEAST (P51862) RHO1 GDP-GTP exchange protein 2
Length = 1356
Score = 43.1 bits (100), Expect = 0.004
Identities = 36/142 (25%), Positives = 59/142 (41%), Gaps = 11/142 (7%)
Query: 156 SMSWCGENICLGIRKAYVIL----NATSGSISEVFTSGRLA-----PPLVVSLPSGELLL 206
S+S+ N+C+G +K + I+ NA + TS A P+ + LL
Sbjct: 1202 SISFLKANLCIGCKKGFQIVSISQNAHESLLDPADTSLEFALRDTLKPMAIYRVGNMFLL 1261
Query: 207 GKDNIGVIVDQNGKLRPEGRIC-WSEAPTEVVIQNPYALALLPRFVEIRSLRGPYPLIQT 265
V+ G + E I W P + I PY LA F+EIR + LI+
Sbjct: 1262 CYTEFAFFVNNQGWRKKESHIIHWEGEPQKFAIWYPYILAFDSNFIEIRKIE-TGELIRC 1320
Query: 266 IVFRNVRHLRQSNNSVIIALEN 287
++ +R L+ S ++ E+
Sbjct: 1321 VLADKIRLLQTSTQEILYCYED 1342
>VP41_YEAST (P38959) Vacuolar assembly protein VPS41 (Vacuolar
morphogenesis protein VAM2)
Length = 992
Score = 39.3 bits (90), Expect = 0.053
Identities = 47/231 (20%), Positives = 99/231 (42%), Gaps = 33/231 (14%)
Query: 536 QEGS---YHVSLVELYKCNSMHREALEIINKSVKESESSQSKIAHRFKP--EAIIEYLKP 590
+EGS Y LV LY + + +A+ + K+ + KI + + I++ +
Sbjct: 666 EEGSNITYRTELVHLYLKENKYTKAIPHLLKAKDLRALTIIKIQNLLPQYLDQIVDIILL 725
Query: 591 LCELDTTLVLEYSMLVLESCPTQTIELFLSGNIPADMVNLY--LKQHAPN----LQATYL 644
+ + + + + S+ +++ + I+L + +Y + P + YL
Sbjct: 726 PYKGEISHISKLSIFEIQTIFNKPIDLLFENRHTISVARIYEIFEHDCPKSFKKILFCYL 785
Query: 645 ELVLSMNEGAVSGTLQNEMVHLYLSEVLDWHADLSSEQKWDEKVYSPKRKKLLSALESIS 704
L ++ + +N+++ LY ++D R+ LL L+ +
Sbjct: 786 IKFLDTDDSFMISPYENQLIELY--------------SEYD-------RQSLLPFLQKHN 824
Query: 705 GYNPEALLKLLPSD-ALYEERAILLGKMNQHELALSLYVHKLHVPELALSY 754
YN E+ +++ S LY E L GK+ + + ALSL + +L P+LA+ +
Sbjct: 825 NYNVESAIEVCSSKLGLYNELIYLWGKIGETKKALSLIIDELKNPQLAIDF 875
>VP41_CAEEL (Q19954) Vacuolar assembly protein VPS41 homolog
Length = 876
Score = 38.5 bits (88), Expect = 0.090
Identities = 33/119 (27%), Positives = 45/119 (37%), Gaps = 24/119 (20%)
Query: 637 PNLQATYLELVLSMNEGAVSGTLQNEMVHLYLSEVLDWHADLSSEQKWDEKVYSPKRKKL 696
P LQ YL ++S NEG ++ V LY ++D+K KL
Sbjct: 616 PKLQLAYLTKLMSRNEGT---EFADKAVQLYA--------------EYDQK-------KL 651
Query: 697 LSALESISGYNPEALLKLLPSDALYEERAILLGKMNQHELALSLYVHKLHVPELALSYC 755
L L + YN KL EE LL K H A+ + V + E + YC
Sbjct: 652 LPFLRKNANYNVNKARKLCSDKGYIEETIYLLAKSGNHYDAVKMMVREYRNMEKVIDYC 710
>GPT2_YEAST (P36148) Glycerol-3-phosphate O-acyltransferase 2 (EC
2.3.1.15) (G-3-P acyltransferase 2) (Dihydroxyacetone
phosphate acyltransferase 2) (EC 2.3.1.42) (DHAP-AT 2)
(Glycerol-3-phosphate / dihydroxyacetone phosphate
acyltransferase 2)
Length = 743
Score = 36.6 bits (83), Expect = 0.34
Identities = 23/64 (35%), Positives = 31/64 (47%), Gaps = 2/64 (3%)
Query: 805 SIRKVTSKSLSRTMSRGSKKIAAIEIAEDAKASQSSDSGRSDADTEEFTEEECTSIMLDE 864
SI + S +LSR SRGS + I I DAK Q G + D +EF E+ +
Sbjct: 654 SIGSLASNALSRVNSRGS--LTDIPIFSDAKQGQWKSEGETSEDEDEFDEKNPAIVQTAR 711
Query: 865 ALDL 868
+ DL
Sbjct: 712 SSDL 715
>MLH1_SCHPO (Q9P7W6) Putative MUTL protein homolog 1 (DNA mismatch
repair protein MLH1)
Length = 684
Score = 36.2 bits (82), Expect = 0.45
Identities = 30/121 (24%), Positives = 57/121 (46%), Gaps = 10/121 (8%)
Query: 742 VHKLHVPELALSYCDHVYESAHKSSVKSLSNIYLMLLQI------YLNPRRTTKNYEKKI 795
VH L+ E+A S CD + E ++ + + M+ I + ++ + YE +
Sbjct: 325 VHFLYDQEIATSICDKLGEILERTDTERSYPLQAMIPSISNTKNAESSSQKAVRTYENYL 384
Query: 796 SNLLSPRNKSIRKVTSKS-LSRTMSRGSKKIAAIEIAEDAKASQSSDSGRSDADTEEFTE 854
PR +SI+ + S + L R+ + +I IE + A +++++ + D TEE E
Sbjct: 385 VR-TDPRERSIKSMLSDNFLQRSSNNYDNEI--IEKVDSANSNKNATNDIKDLQTEEIVE 441
Query: 855 E 855
E
Sbjct: 442 E 442
>KNTC_HUMAN (P50748) Kinetochore-associated protein 1 (Rough deal
homolog) (hRod) (HsROD) (Rod)
Length = 2209
Score = 35.8 bits (81), Expect = 0.58
Identities = 82/416 (19%), Positives = 164/416 (38%), Gaps = 75/416 (18%)
Query: 577 HRFKPEAIIEYLKPLCELDTTLVLEYSMLVLESCPTQTIELFL--------SGNIPADMV 628
H + +I+ L ++DT+L+LEY C ++LF+ +G D
Sbjct: 1425 HFLTKKDLIKALVENIDMDTSLILEYCSTFQLDCDA-VLQLFIETLLHNTNAGQGQGDAS 1483
Query: 629 NLYLKQHAPNLQATYLELVLSMNEG-----AVSGTLQN------EMVHLYLSEVLDWHAD 677
K+ P L A LE+V + ++SG L EM+ + L +
Sbjct: 1484 MDSAKRRHPKLLAKALEMVPLLTSTKDLVISLSGILHKLDPYDYEMIEVVLKVI------ 1537
Query: 678 LSSEQKWDEKVYSPKRKKLLSALESISGYN--------------------PEALLKLLPS 717
++ DEK+ + + LS L+ + Y P A LP
Sbjct: 1538 ----ERADEKITNININQALSILKHLKSYRRISPPVDLEYQYMLEHVITLPSAAQTRLPF 1593
Query: 718 DALYEERA-----ILLGKMNQHELALSLYVHKL---HVPELALSYCDHVYESAHKSSVKS 769
++ A IL ++++ L + KL + L +S HV+E K +
Sbjct: 1594 HLIFFGTAQNFWKILSTELSEESFPTLLLISKLMKFSLDTLYVSTAKHVFEKKLKPKLLK 1653
Query: 770 LSNIYLMLLQIYLNPRRTTKNYEKKISNLLSPRNKSIRKVTSKSLSRTMSRGSKKIAAIE 829
L+ L +N + TK + S LLS N + SL++ + GS KI+A++
Sbjct: 1654 LTQAKSSTL---IN-KEITKITQTIESCLLSIVNPEWAVAIAISLAQDIPEGSFKISALK 1709
Query: 830 IAEDAKASQSSDSGRSDADTEEFTEEECTSIMLDEALDLLSRRWDRINGAQALKLLPKET 889
++ + +++ E+ +++ + L + RR +G +A+ L+ +
Sbjct: 1710 F-----CLYLAERWLQNIPSQDEKREKAEALL--KKLHIQYRR----SGTEAV-LIAHKL 1757
Query: 890 KLQNLLPILGPLVRKSSEMYRNCSVVRSLRQSENLQVKDELYNKRKAVIKISDDNM 945
+ L ++G +Y + S+ + ++ S D ++ K + ++++ N+
Sbjct: 1758 NTEEYLRVIGKPAHLIVSLYEHPSINQRIQNSSGTDYPD-IHAAAKEIAEVNEINL 1812
>TNIK_MOUSE (P83510) Traf2 and NCK interacting kinase (EC 2.7.1.37)
(Fragments)
Length = 916
Score = 35.0 bits (79), Expect = 0.99
Identities = 24/101 (23%), Positives = 49/101 (47%), Gaps = 8/101 (7%)
Query: 164 ICLGIRKAYVILNATSGSISEVFT----SGRLAPPLVVSLPSG---ELLLGKDNIGVIVD 216
+ G + +++ SG+ +++ G + P +V LP E+L+ ++ GV V+
Sbjct: 763 VIFGSHTGFHVIDVDSGNSYDIYIPSHIQGNITPHAIVILPKTDGMEMLVCYEDEGVYVN 822
Query: 217 QNGKLRPEGRICWSEAPTEVV-IQNPYALALLPRFVEIRSL 256
G++ + + W E PT V I + + + +EIRS+
Sbjct: 823 TYGRITKDVVLQWGEMPTSVAYIHSNQIMGWGEKAIEIRSV 863
>TNIK_HUMAN (Q9UKE5) TRAF2 and NCK interacting kinase (EC 2.7.1.37)
Length = 1360
Score = 35.0 bits (79), Expect = 0.99
Identities = 24/101 (23%), Positives = 49/101 (47%), Gaps = 8/101 (7%)
Query: 164 ICLGIRKAYVILNATSGSISEVFT----SGRLAPPLVVSLPSG---ELLLGKDNIGVIVD 216
+ G + +++ SG+ +++ G + P +V LP E+L+ ++ GV V+
Sbjct: 1207 VIFGSHTGFHVIDVDSGNSYDIYIPSHIQGNITPHAIVILPKTDGMEMLVCYEDEGVYVN 1266
Query: 217 QNGKLRPEGRICWSEAPTEVV-IQNPYALALLPRFVEIRSL 256
G++ + + W E PT V I + + + +EIRS+
Sbjct: 1267 TYGRITKDVVLQWGEMPTSVAYIHSNQIMGWGEKAIEIRSV 1307
>ROM1_YEAST (P53046) RHO1 GDP-GTP exchange protein 1 (Protein kinase C
suppressor SKC1)
Length = 1155
Score = 35.0 bits (79), Expect = 0.99
Identities = 40/181 (22%), Positives = 72/181 (39%), Gaps = 38/181 (20%)
Query: 147 DFGVLDVVKSMSWCGENICLGIRKAYVILNATSGSISEVFTSGRLAPPLVVSLPSGELLL 206
DF V S S+ IC+G +K ILN + EV +++L ++L
Sbjct: 987 DFSVDSEPLSFSFLENKICIGCKKNIKILN-----VPEVCDKNGFKMRELLNLHDNKVLA 1041
Query: 207 G--KDNIGVI-------------------VDQNGKLRPEGRIC--WSEAPTEVVIQNPYA 243
K+ V+ +++ GK R E + C W P + PY
Sbjct: 1042 NMYKETFKVVSMFPIKNSTFACFPELCFFLNKQGK-REETKGCFHWEGEPEQFACSYPYI 1100
Query: 244 LALLPRFVEIRSLRGPYPLIQTIVFRNVRHLRQSNNSVIIALENSIHCLFPVPLGAQIVQ 303
+A+ F+EIR + L++ ++ +R L+ ++ E+ P G +I++
Sbjct: 1101 VAINSNFIEIRHIENG-ELVRCVLGNKIRMLKSYAKKILYCYED--------PQGFEIIE 1151
Query: 304 L 304
L
Sbjct: 1152 L 1152
>PKND_CHLMU (Q9PK92) Serine/threonine-protein kinase pknD (EC
2.7.1.37)
Length = 934
Score = 34.7 bits (78), Expect = 1.3
Identities = 27/109 (24%), Positives = 48/109 (43%), Gaps = 4/109 (3%)
Query: 697 LSALESISGYNPEALLKLLPSDALYEERAILLGKMNQHELALSLYVHKLHVPELALSYCD 756
LS + + G + A L+ L +Y+ + ++ LAL Y +P L C
Sbjct: 541 LSTFDLLHG-STGAPLEYLGKALVYQRNGSFVEEIRSLLLALKRYPQHPEIPRLKDHLCF 599
Query: 757 HVYESAHKSSVKSLSNIYLMLLQIYLNPRRTTKNYEKKISNLLSPRNKS 805
+Y+S HK ++L MLL +++ P + E++ L R +S
Sbjct: 600 RLYDSLHKHRSEAL---VFMLLILWIAPEKIGLREEERFLEFLHHRQQS 645
>NAK1_SCHPO (O75011) Serine/threonine-protein kinase nak1 (EC
2.7.1.37) (N-rich kinase 1)
Length = 652
Score = 34.3 bits (77), Expect = 1.7
Identities = 24/91 (26%), Positives = 42/91 (45%), Gaps = 2/91 (2%)
Query: 371 TSIILPKTTIVHDSDKLDIFGDPLHLSRGSSMSDDMEPSSASNMSELDDNAELESKKMSH 430
T ++PK++ VH+ + L L + S +SDD PS+A S D E+ ++
Sbjct: 354 TGTVIPKSSTVHEPPSSNDSHPLLQLFKDSKISDDDSPSNAEGASTEDSKGEVSYSQIEL 413
Query: 431 NMLMALIKFLHKKRHSIIEKATAEGTEEVVF 461
L + L K+ +I K T + + +F
Sbjct: 414 PSLDS--SNLSSKKSTIQSKHTKQAEDYDLF 442
>RECN_AQUAE (O66834) DNA repair protein recN (Recombination protein
N)
Length = 520
Score = 33.9 bits (76), Expect = 2.2
Identities = 21/53 (39%), Positives = 32/53 (59%), Gaps = 4/53 (7%)
Query: 647 VLSMNEGAVSGTLQNEMVHLYLSE----VLDWHADLSSEQKWDEKVYSPKRKK 695
VL + EG +S QNE ++L+ + +LD A+L+ + K EKVY+ RKK
Sbjct: 93 VLELLEGKISLQGQNEFINLFREDFQRKLLDTFANLNDKVKELEKVYNSLRKK 145
>G59F_DROME (Q9W1N5) Putative gustatory receptor 59f
Length = 406
Score = 33.5 bits (75), Expect = 2.9
Identities = 21/71 (29%), Positives = 37/71 (51%), Gaps = 2/71 (2%)
Query: 739 SLYVHKLHVPELALSYCDHVYESAHKSSVKSLSNIYLMLLQIYLNPRRTTKNYEKKISNL 798
+++ HK V LS +V + S S + YL+L I RR T+ E+++++L
Sbjct: 171 NIWTHKFVVYRSILSINSYVMPNIISSI--SFAQYYLLLQGIAWRQRRLTEGLERELTHL 228
Query: 799 LSPRNKSIRKV 809
SPR ++K+
Sbjct: 229 HSPRISEVQKI 239
>Y111_ENCCU (Q8STB5) Hypothetical protein
ECU01_0110/ECU01_1500/ECU08_0040
Length = 592
Score = 33.1 bits (74), Expect = 3.8
Identities = 28/113 (24%), Positives = 51/113 (44%), Gaps = 5/113 (4%)
Query: 762 AHKSSVKSLSNIYLMLLQIYLNPRRTTKNYEKKISNLLSPRNKSIRKVTSKSLSRTMSRG 821
AHK ++ +M + + + K Y K++N++ R + + K++ M R
Sbjct: 266 AHKEHGGDVTKELVMQMLLGKKGKNIDKRYINKVANVVKERQRRREREIEKNMKELM-RD 324
Query: 822 SKKIAAIEIA---EDAKASQSSDS-GRSDADTEEFTEEECTSIMLDEALDLLS 870
+K + E A E AK+ + +S G S+A EE E E + E + + S
Sbjct: 325 EEKAKSKEKAKSKEKAKSKKKGESVGVSEAKEEEKKESETEEVEASEEVGIPS 377
>SL2B_HUMAN (Q8NEV8) Slp homolog lacking C2 domains-b (Exophilin 5)
Length = 1989
Score = 33.1 bits (74), Expect = 3.8
Identities = 62/259 (23%), Positives = 90/259 (33%), Gaps = 48/259 (18%)
Query: 747 VPELALSYCDHVYESAHKSSVKSLSNIYLMLLQIYLNPRRTTKNYEKKISNLLSPRNKSI 806
+P + S+ H +S K SL N QI N NY + N SP +
Sbjct: 1022 LPRKSSSFLIHGRQSGSKIMAASLRN-GPPPFQIKNNVEDAMGNY---MLNKFSPSSPES 1077
Query: 807 RKVTSKSLS--------------RTMSRGSKKIAAIEIAEDAKASQSSDSGRSDADT-EE 851
SK LS S GS + + + S SG A T E
Sbjct: 1078 ANECSKVLSDSALEAPEATERMTNVKSSGSTSVRKGPLPFLINRAMSCPSGEPHASTGRE 1137
Query: 852 FTEEECTSIMLDEALDLLSRRWDRINGAQALKLLPKETKLQNLLPILGPLVRKSSEMYRN 911
++ TS M +A +L R W+RI + P+ SS R+
Sbjct: 1138 GRKKPLTSGM--DASELTPRAWERI---------------------ISPVESDSS--VRD 1172
Query: 912 CSVVRSLRQSENLQVKDELYNK----RKAVIKISDDNMCSLCHKKIGTSVFAVYPNGKTL 967
CS+ + Q EN Q E K R++V +S+++ C G KT
Sbjct: 1173 CSLTKRQHQKENFQEYTEKEGKMAASRRSVFALSNEDPLPFCSDLSGKERGKTLHKVKTT 1232
Query: 968 VHFVCFRDSQSMKAVAKVS 986
F D ++K + VS
Sbjct: 1233 STFSVSGDEDNVKCLEVVS 1251
>MI15_CAEEL (Q23356) Serine/threonine-protein kinase mig-15 (EC
2.7.1.37) (Abnormal cell migration protein 15)
Length = 1080
Score = 33.1 bits (74), Expect = 3.8
Identities = 27/101 (26%), Positives = 49/101 (47%), Gaps = 8/101 (7%)
Query: 164 ICLGIRKAYVILNATSGSISEVFT---SGRLAPP-LVVSLPSG---ELLLGKDNIGVIVD 216
+ G + ++ S ++ +++T SG+ P +V LP+ +LLL DN GV V+
Sbjct: 927 VLYGSTGGFHAIDLDSAAVYDIYTPAQSGQTTTPHCIVVLPNSNGMQLLLCYDNEGVYVN 986
Query: 217 QNGKLRPEGRICWSEAPTEVV-IQNPYALALLPRFVEIRSL 256
G++ + W E P+ V I + + +EIRS+
Sbjct: 987 TYGRMTKNVVLQWGEMPSSVAYISTGQIMGWGNKAIEIRSV 1027
>MAD1_DEBHA (Q6BSY1) Spindle assembly checkpoint component MAD1
(Mitotic arrest deficient protein 1)
Length = 669
Score = 33.1 bits (74), Expect = 3.8
Identities = 62/306 (20%), Positives = 117/306 (37%), Gaps = 38/306 (12%)
Query: 466 NNFESYNSNRFKKINKRHGSIPVSSEAREMASILDTALLQAMLLTGQPSMAENLLRVLNY 525
+N + S K+ + ++ +E +D Q +L + +LL+ L
Sbjct: 321 SNMQQKYSEAISKVESLNETVSTKTE------FIDKLERQKLLNVKEIEYLRDLLKKLEE 374
Query: 526 CDLKICEEILQEGSYHVSLVELYKCNSMHREALEIINKSVKESE-----SSQSK------ 574
+LK +E + S + L K +R + + K + E S SK
Sbjct: 375 LNLKRSKEETNDKSTEQYMTNLEKLVDEYRTEINTLQKQMLSQEIQSNVQSGSKRPRIID 434
Query: 575 --IAHRFKPEAIIEYLKPLCELDTTLVLEYSMLVLESCPTQTIELFLSGNIPADMVNLYL 632
I + FK + + + L L T LEY+ L Q +E N ++ L L
Sbjct: 435 NGIQNDFKSQVSVLEKENLKLLSTIKNLEYNNKTLGE-KLQNLESL--DNKKKELHILQL 491
Query: 633 KQHAPN----LQATYLELVLSMNEGAVSGTLQNEMVHLYLSEVLDWHAD---LSSEQKWD 685
K + + ++ L+L+ N+ + ++N+ H + + L + L + K D
Sbjct: 492 KSNPASQDQLIKQQTLDLLSKENQEMIETFVKNKSAHDMIPKSLFERQENDKLQLQTKID 551
Query: 686 E---------KVYSPKRKKLLSALESISGYNPEALLKLLPSDALYEERAILLGKMNQHEL 736
+ +YS K + +LS + GY E L + S+ L ++ MN +
Sbjct: 552 QLNKRISRLRDIYSQKSRDILSVISKFFGYTIEFLPSPINSNDLSSRIKLVSKYMNNKDE 611
Query: 737 ALSLYV 742
+ S Y+
Sbjct: 612 SNSAYL 617
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 106,899,572
Number of Sequences: 164201
Number of extensions: 4398904
Number of successful extensions: 12481
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 12442
Number of HSP's gapped (non-prelim): 47
length of query: 992
length of database: 59,974,054
effective HSP length: 120
effective length of query: 872
effective length of database: 40,269,934
effective search space: 35115382448
effective search space used: 35115382448
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC147964.8


