

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147964.4 + phase: 0 /pseudo
(276 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
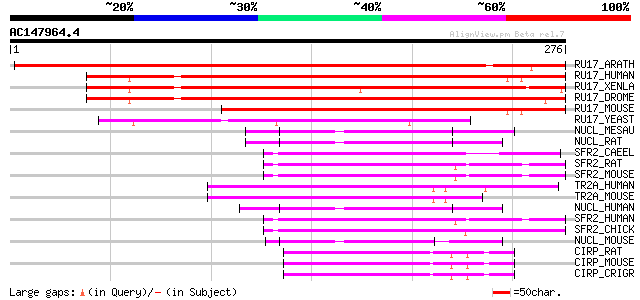
Score E
Sequences producing significant alignments: (bits) Value
RU17_ARATH (Q42404) U1 small nuclear ribonucleoprotein 70 kDa (U... 433 e-121
RU17_HUMAN (P08621) U1 small nuclear ribonucleoprotein 70 kDa (U... 250 2e-66
RU17_XENLA (P09406) U1 small nuclear ribonucleoprotein 70 kDa (U... 238 1e-62
RU17_DROME (P17133) U1 small nuclear ribonucleoprotein 70 kDa (U... 219 7e-57
RU17_MOUSE (Q62376) U1 small nuclear ribonucleoprotein 70 kDa (U... 213 3e-55
RU17_YEAST (Q00916) U1 small nuclear ribonucleoprotein 70 kDa ho... 119 8e-27
NUCL_MESAU (P08199) Nucleolin (Protein C23) 70 6e-12
NUCL_RAT (P13383) Nucleolin (Protein C23) 70 7e-12
SFR2_CAEEL (Q09511) Probable splicing factor, arginine/serine-ri... 69 1e-11
SFR2_RAT (Q6PDU1) Splicing factor, arginine/serine-rich 2 (Splic... 66 1e-10
SFR2_MOUSE (Q62093) Splicing factor, arginine/serine-rich 2 (Spl... 66 1e-10
TR2A_HUMAN (Q13595) Transformer-2 protein homolog (TRA-2 alpha) 64 3e-10
TR2A_MOUSE (Q6PFR5) Transformer-2 protein homolog (TRA-2 alpha) 64 4e-10
NUCL_HUMAN (P19338) Nucleolin (Protein C23) 64 5e-10
SFR2_HUMAN (Q01130) Splicing factor, arginine/serine-rich 2 (Spl... 63 9e-10
SFR2_CHICK (P30352) Splicing factor, arginine/serine-rich 2 (Spl... 62 1e-09
NUCL_MOUSE (P09405) Nucleolin (Protein C23) 62 2e-09
CIRP_RAT (P60825) Cold-inducible RNA-binding protein (Glycine-ri... 62 2e-09
CIRP_MOUSE (P60824) Cold-inducible RNA-binding protein (Glycine-... 62 2e-09
CIRP_CRIGR (P60826) Cold-inducible RNA-binding protein (Glycine-... 62 2e-09
>RU17_ARATH (Q42404) U1 small nuclear ribonucleoprotein 70 kDa (U1
snRNP 70 kDa) (snRNP70) (U1-70K)
Length = 427
Score = 433 bits (1113), Expect = e-121
Identities = 212/275 (77%), Positives = 238/275 (86%), Gaps = 4/275 (1%)
Query: 3 DPLLRNQNAAVQARTKAQNRANVLQLKLIGQSHPTGLTANLLKLFEPRPPLEYKPPPEKR 62
DP LRN NAAVQAR K QNRANVLQLKL+GQSHPTGLT NLLKLFEPRPPLEYKPPPEKR
Sbjct: 6 DPFLRNPNAAVQARAKVQNRANVLQLKLMGQSHPTGLTNNLLKLFEPRPPLEYKPPPEKR 65
Query: 63 KCPPLTGMAQFVSKFAEPGEPEYSPPVPVVETPAERRARVHKLRLEKGAAKAAEELEKYD 122
KCPP TGMAQFVS FAEPG+PEY+PP P VE P+++R R+HKLRLEKG KAAE+L+KYD
Sbjct: 66 KCPPYTGMAQFVSNFAEPGDPEYAPPKPEVELPSQKRERIHKLRLEKGVEKAAEDLKKYD 125
Query: 123 PHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIE 182
P+NDPN +GDPYKTLFV+RL+YE++ES+IKREFESYG IKRV LVTD +NKP+GYAFIE
Sbjct: 126 PNNDPNATGDPYKTLFVSRLNYESSESKIKREFESYGPIKRVHLVTDQLTNKPKGYAFIE 185
Query: 183 YLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRH 242
Y+HTRDMKAAYKQADG+KI+GRRVLVDVERGRTVPNWRPRRLGGGLGT+RVGG E
Sbjct: 186 YMHTRDMKAAYKQADGQKIDGRRVLVDVERGRTVPNWRPRRLGGGLGTSRVGGGE---EI 242
Query: 243 SGREQQQSGPSRSEEP-RVREERHADRDREKSRER 276
G +Q Q S+SEEP R REER R++ K RER
Sbjct: 243 VGEQQPQGRTSQSEEPSRPREEREKSREKGKERER 277
>RU17_HUMAN (P08621) U1 small nuclear ribonucleoprotein 70 kDa (U1
snRNP 70 kDa) (snRNP70) (U1-70K)
Length = 437
Score = 250 bits (639), Expect = 2e-66
Identities = 132/253 (52%), Positives = 166/253 (65%), Gaps = 18/253 (7%)
Query: 39 LTANLLKLFEPRPPLEYKPP-----PEKRKCPPLTGMAQFVSKFAEPGEPEYSPPVPVVE 93
L NLL LF PR P+ Y PP EK P G+A ++ +F +P + +PP E
Sbjct: 5 LPPNLLALFAPRDPIPYLPPLEKLPHEKHHNQPYCGIAPYIREFEDPRD---APPPTRAE 61
Query: 94 TPAERRARVHKLRLEKGAAKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKR 153
T ER R + ++E+ + EL+ +DPHNDPN GD +KTLFVAR++Y+TTES+++R
Sbjct: 62 TREERMERKRREKIERRQQEVETELKMWDPHNDPNAQGDAFKTLFVARVNYDTTESKLRR 121
Query: 154 EFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERG 213
EFE YG IKR+ +V S KPRGYAFIEY H RDM +AYK ADG+KI+GRRVLVDVERG
Sbjct: 122 EFEVYGPIKRIHMVYSKRSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRRVLVDVERG 181
Query: 214 RTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGRE-----QQQSGPS-----RSEEPRVREE 263
RTV WRPRRLGGGLG TR GG +VN RHSGR+ ++ GPS + R RE
Sbjct: 182 RTVKGWRPRRLGGGLGGTRRGGADVNIRHSGRDDTSRYDERPGPSPLPHRDRDRDRERER 241
Query: 264 RHADRDREKSRER 276
R R+R+K RER
Sbjct: 242 RERSRERDKERER 254
>RU17_XENLA (P09406) U1 small nuclear ribonucleoprotein 70 kDa (U1
snRNP 70 kDa) (snRNP70) (U1-70K)
Length = 471
Score = 238 bits (607), Expect = 1e-62
Identities = 130/247 (52%), Positives = 161/247 (64%), Gaps = 13/247 (5%)
Query: 39 LTANLLKLFEPRPPLEYKPP-----PEKRKCPPLTGMAQFVSKFAEPGEPEYSPPVPVVE 93
L NLL LF PR P+ Y PP EK P G+A ++ +F +P + +PP E
Sbjct: 5 LPPNLLALFAPRDPVPYLPPLDKLPHEKHHNQPYCGIAPYIREFEDPRD---APPPTRAE 61
Query: 94 TPAERRARVHKLRLEKGAAKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKR 153
T ER R + ++E+ EL+ +DPHND N GD +KTLFVAR++Y+TTES+++R
Sbjct: 62 TREERMERKRREKIERRQQDVENELKIWDPHNDQNAQGDAFKTLFVARVNYDTTESKLRR 121
Query: 154 EFESYGAIKRVRLVTDAESN---KPRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDV 210
EFE YG IKR+ +V + S KPRGYAFIEY H RDM +AYK ADG+KI+GRRVLVDV
Sbjct: 122 EFEVYGPIKRIHIVYNKGSEGSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRRVLVDV 181
Query: 211 ERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGREQQQSGPSRSEEPRVREERHADRDR 270
ERGRTV WRPRRLGGGLG TR GG +VN RHSGR+ R E R R+ R R+R
Sbjct: 182 ERGRTVKGWRPRRLGGGLGGTRRGGADVNIRHSGRDDTSRYDERDRE-RERDRRERSRER 240
Query: 271 EKS-RER 276
EK RER
Sbjct: 241 EKEPRER 247
>RU17_DROME (P17133) U1 small nuclear ribonucleoprotein 70 kDa (U1
snRNP 70 kDa) (snRNP70)
Length = 448
Score = 219 bits (557), Expect = 7e-57
Identities = 120/262 (45%), Positives = 161/262 (60%), Gaps = 27/262 (10%)
Query: 39 LTANLLKLFEPRPPLEYKPP----PEKRKCPPLTGMAQFVSKFAEPGEPEYSPPVPVVET 94
L NLL LF R P+ + PP P ++K G+A+F++ F +P + +P VET
Sbjct: 5 LPPNLLALFAAREPIPFMPPVDKLPHEKKSRGYLGVAKFMADFEDPKD---TPLPKTVET 61
Query: 95 PAERRARVHKLRLEKGAAKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKRE 154
ER R + + E+ A K E+ +DP N + DP++TLF+AR++Y+T+ES+++RE
Sbjct: 62 RQERLERRRREKAEQVAYKLEREIALWDPTEIKNATEDPFRTLFIARINYDTSESKLRRE 121
Query: 155 FESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGR 214
FE YG IK++ L+ D ES KP+GYAFIEY H RDM AAYK ADG+KI+ +RVLVDVER R
Sbjct: 122 FEFYGPIKKIVLIHDQESGKPKGYAFIEYEHERDMHAAYKHADGKKIDSKRVLVDVERAR 181
Query: 215 TVPNWRPRRLGGGLGTTRVGGDEVNQRHSGREQQQSGPSRSEEPRVREERH--------- 265
TV W PRRLGGGLG TR GG++VN +HSGRE + R R RE+R
Sbjct: 182 TVKGWLPRRLGGGLGGTRRGGNDVNIKHSGREDNERERERYRLEREREDREGPGRGGGSN 241
Query: 266 -----------ADRDREKSRER 276
A+R R +SRER
Sbjct: 242 GLDARPGRGFGAERRRSRSRER 263
Score = 30.0 bits (66), Expect = 6.4
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 6/83 (7%)
Query: 194 KQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGREQQQSGPS 253
++ D + GR + R R+ R +R G +R DE ++R R ++
Sbjct: 262 ERRDRERDRGRGAVASNGRSRSRSRERRKRRAG----SRERYDEFDRRD--RRDRERERD 315
Query: 254 RSEEPRVREERHADRDREKSRER 276
R E +++R R+RE SRER
Sbjct: 316 RDREREKKKKRSKSRERESSRER 338
>RU17_MOUSE (Q62376) U1 small nuclear ribonucleoprotein 70 kDa (U1
SNRNP 70 kDa) (snRNP70) (Fragment)
Length = 378
Score = 213 bits (543), Expect = 3e-55
Identities = 106/181 (58%), Positives = 131/181 (71%), Gaps = 10/181 (5%)
Query: 106 RLEKGAAKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVR 165
++E+ + EL+ +DPHNDPN GD +KTLFVAR++Y+TTES+++REFE YG IKR+
Sbjct: 4 KIERRQQEVETELKMWDPHNDPNAQGDAFKTLFVARVNYDTTESKLRREFEVYGPIKRIH 63
Query: 166 LVTDAESNKPRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLG 225
+V S KPRGYAFIEY H RDM +AYK ADG+KI+GRRVLVDVERGRTV WRPRRLG
Sbjct: 64 MVYSKRSGKPRGYAFIEYEHERDMHSAYKHADGKKIDGRRVLVDVERGRTVKGWRPRRLG 123
Query: 226 GGLGTTRVGGDEVNQRHSGRE-----QQQSGPS-----RSEEPRVREERHADRDREKSRE 275
GGLG TR GG +VN RHSGR+ ++ GPS + R RE R R+R+K RE
Sbjct: 124 GGLGGTRRGGADVNIRHSGRDDTSRYDERPGPSPLPHRDRDRDRERERRERSRERDKERE 183
Query: 276 R 276
R
Sbjct: 184 R 184
>RU17_YEAST (Q00916) U1 small nuclear ribonucleoprotein 70 kDa
homolog
Length = 300
Score = 119 bits (298), Expect = 8e-27
Identities = 69/194 (35%), Positives = 110/194 (56%), Gaps = 12/194 (6%)
Query: 45 KLFEPRPPLEYKPPPE-----KRKCPPLTGMAQFVSKFAEPGEPEYSPPVPVVETPAERR 99
+LF+PRPPL YK P + ++ P +TG+A +S + E+ P
Sbjct: 14 RLFKPRPPLSYKRPTDYPYAKRQTNPNITGVANLLSTSLKHYMEEFPEGSPNNHLQRYED 73
Query: 100 ARVHKLRLEKGAAKAAEELEKYDPHNDPNISG-DPYKTLFVARLSYETTESRIKREFESY 158
++ K+ K A L+ ++P+ DP+I DPY+T+F+ RL Y+ E +++ F +
Sbjct: 74 IKLSKI---KNAQLLDRRLQNWNPNVDPHIKDTDPYRTIFIGRLPYDLDEIELQKYFVKF 130
Query: 159 GAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQAD---GRKIEGRRVLVDVERGRT 215
G I+++R+V D + K +GYAFI + K A+K+ G +I+ R +VD+ERGRT
Sbjct: 131 GEIEKIRIVKDKITQKSKGYAFIVFKDPISSKMAFKEIGVHRGIQIKDRICIVDIERGRT 190
Query: 216 VPNWRPRRLGGGLG 229
V ++PRRLGGGLG
Sbjct: 191 VKYFKPRRLGGGLG 204
>NUCL_MESAU (P08199) Nucleolin (Protein C23)
Length = 713
Score = 70.1 bits (170), Expect = 6e-12
Identities = 45/134 (33%), Positives = 64/134 (47%), Gaps = 12/134 (8%)
Query: 118 LEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRG 177
LE P PN P KTLFV LS +TTE +K FE G++ R R+VTD E+ +G
Sbjct: 554 LELQGPRGSPNARSQPSKTLFVKGLSEDTTEETLKESFE--GSV-RARIVTDRETGSSKG 610
Query: 178 YAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDE 237
+ F+++ D KAA + + +I+G +V +D W + GG G G
Sbjct: 611 FGFVDFNSEEDAKAAKEAMEDGEIDGNKVTLD---------WAKPKGEGGFGGRGGGRGG 661
Query: 238 VNQRHSGREQQQSG 251
R GR + G
Sbjct: 662 FGGRGGGRGGGRGG 675
Score = 53.9 bits (128), Expect = 4e-07
Identities = 31/86 (36%), Positives = 45/86 (52%), Gaps = 4/86 (4%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
KTL ++ LSY TE ++ FE IK V + K +GYAFIE+ D K A
Sbjct: 485 KTLVLSNLSYSATEETLQEVFEKATFIK----VPQNQQGKSKGYAFIEFASFEDAKEALN 540
Query: 195 QADGRKIEGRRVLVDVERGRTVPNWR 220
+ +IEGR + ++++ R PN R
Sbjct: 541 SCNKMEIEGRTIRLELQGPRGSPNAR 566
Score = 39.3 bits (90), Expect = 0.011
Identities = 23/72 (31%), Positives = 37/72 (50%), Gaps = 5/72 (6%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
+TL LS+ TE +K FE +RLV+ + K +G A+IE+ D + +
Sbjct: 393 RTLLAKNLSFNITEDELKEVFED---ALEIRLVS--QDGKSKGIAYIEFKSEADAEKNLE 447
Query: 195 QADGRKIEGRRV 206
+ G +I+GR V
Sbjct: 448 EKQGAEIDGRSV 459
>NUCL_RAT (P13383) Nucleolin (Protein C23)
Length = 712
Score = 69.7 bits (169), Expect = 7e-12
Identities = 44/128 (34%), Positives = 62/128 (48%), Gaps = 12/128 (9%)
Query: 118 LEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRG 177
LE P PN P KTLFV LS +TTE +K FE G++ R R+VTD E+ +G
Sbjct: 557 LELQGPRGSPNARSQPSKTLFVKGLSEDTTEETLKESFE--GSV-RARIVTDRETGSSKG 613
Query: 178 YAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDE 237
+ F+++ D KAA + + +I+G +V +D W + GG G G
Sbjct: 614 FGFVDFNSEEDAKAAKEAMEDGEIDGNKVTLD---------WAKPKGEGGFGGRGGGRGG 664
Query: 238 VNQRHSGR 245
R GR
Sbjct: 665 FGGRGGGR 672
Score = 52.4 bits (124), Expect = 1e-06
Identities = 31/86 (36%), Positives = 44/86 (51%), Gaps = 4/86 (4%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
KTL ++ LSY TE ++ FE IK V K +GYAFIE+ D K A
Sbjct: 488 KTLVLSNLSYSATEETLQEVFEKATFIK----VPQNPHGKSKGYAFIEFASFEDAKEALN 543
Query: 195 QADGRKIEGRRVLVDVERGRTVPNWR 220
+ +IEGR + ++++ R PN R
Sbjct: 544 SCNKMEIEGRTIRLELQGPRGSPNAR 569
Score = 38.1 bits (87), Expect = 0.024
Identities = 21/72 (29%), Positives = 36/72 (49%), Gaps = 5/72 (6%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
+TL LS+ TE +K FE I+ V ++ + +G A+IE+ D + +
Sbjct: 396 RTLLAKNLSFNITEDELKEVFEDAVEIRLV-----SQDGRSKGIAYIEFKSEADAEKNLE 450
Query: 195 QADGRKIEGRRV 206
+ G +I+GR V
Sbjct: 451 EKQGAEIDGRSV 462
>SFR2_CAEEL (Q09511) Probable splicing factor, arginine/serine-rich
2 (Splicing factor SC35) (SC-35) (Splicing component, 35
kDa)
Length = 196
Score = 69.3 bits (168), Expect = 1e-11
Identities = 44/148 (29%), Positives = 71/148 (47%), Gaps = 18/148 (12%)
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P+I+G +L + LSY+TT + ++R FE YG I V + D S + +G+ F+ +
Sbjct: 13 PDING--LTSLKIDNLSYQTTPNDLRRTFERYGDIGDVHIPRDKYSRQSKGFGFVRFYER 70
Query: 187 RDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGRE 246
RD + A + DG+ ++GR + V + + + R R GGG GR
Sbjct: 71 RDAEHALDRTDGKLVDGRELRVTLAKYDRPSDERGGRGGGG----------------GRR 114
Query: 247 QQQSGPSRSEEPRVREERHADRDREKSR 274
+ +S RS PR R R R ++R
Sbjct: 115 RSRSPRRRSRSPRYSRSRSPRRSRSRTR 142
>SFR2_RAT (Q6PDU1) Splicing factor, arginine/serine-rich 2 (Splicing
factor SC35) (SC-35) (Splicing component, 35 kDa)
Length = 220
Score = 65.9 bits (159), Expect = 1e-10
Identities = 49/157 (31%), Positives = 72/157 (45%), Gaps = 13/157 (8%)
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P++ G +L V L+Y T+ ++R FE YG + V + D + + RG+AF+ +
Sbjct: 7 PDVEG--MTSLKVDNLTYRTSPDTLRRVFEKYGRVGDVYIPRDRYTKESRGFAFVRFHDK 64
Query: 187 RDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWR-------PRRLGGGLGTTRVGGDEVN 239
RD + A DG ++GR + V + R P+ PRR GGG G R
Sbjct: 65 RDAEDAMDAMDGAVLDGRELRVQMARYGRPPDSHHSRRGPPPRRYGGG-GYGRRSRSPRR 123
Query: 240 QRHSGREQQQSGPSRSEEPRVREERHADRDREKSRER 276
+R S + SRS R R R R R +SR R
Sbjct: 124 RRRSRSRSRSRSRSRS---RSRYSRSKSRSRTRSRSR 157
>SFR2_MOUSE (Q62093) Splicing factor, arginine/serine-rich 2
(Splicing factor SC35) (SC-35) (Splicing component, 35
kDa) (PR264 protein)
Length = 220
Score = 65.9 bits (159), Expect = 1e-10
Identities = 49/157 (31%), Positives = 72/157 (45%), Gaps = 13/157 (8%)
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P++ G +L V L+Y T+ ++R FE YG + V + D + + RG+AF+ +
Sbjct: 7 PDVEG--MTSLKVDNLTYRTSPDTLRRVFEKYGRVGDVYIPRDRYTKESRGFAFVRFHDK 64
Query: 187 RDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWR-------PRRLGGGLGTTRVGGDEVN 239
RD + A DG ++GR + V + R P+ PRR GGG G R
Sbjct: 65 RDAEDAMDAMDGAVLDGRELRVQMARYGRPPDSHHSRRGPPPRRYGGG-GYGRRSRSPRR 123
Query: 240 QRHSGREQQQSGPSRSEEPRVREERHADRDREKSRER 276
+R S + SRS R R R R R +SR R
Sbjct: 124 RRRSRSRSRSRSRSRS---RSRYSRSKSRSRTRSRSR 157
>TR2A_HUMAN (Q13595) Transformer-2 protein homolog (TRA-2 alpha)
Length = 282
Score = 64.3 bits (155), Expect = 3e-10
Identities = 51/194 (26%), Positives = 80/194 (40%), Gaps = 19/194 (9%)
Query: 99 RARVHKLRLEKGAAKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESY 158
R+R + + +++ + H + DP L V LS TTE ++ F Y
Sbjct: 83 RSRSYTPEYRRRRSRSHSPMSNRRRHTGSRANPDPNTCLGVFGLSLYTTERDLREVFSRY 142
Query: 159 GAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVD---VERGRT 215
G + V +V D + + RG+AF+ + D K A ++A+G +++GRR+ VD +R T
Sbjct: 143 GPLSGVNVVYDQRTGRSRGFAFVYFERIDDSKEAMERANGMELDGRRIRVDYSITKRAHT 202
Query: 216 ----VPNWRPRRLGGGLGTTRVGG------------DEVNQRHSGREQQQSGPSRSEEPR 259
+ RP GGG G GG D R R + R P
Sbjct: 203 PTPGIYMGRPTHSGGGGGGGGGGGGGGGGRRRDSYYDRGYDRGYDRYEDYDYRYRRRSPS 262
Query: 260 VREERHADRDREKS 273
R+ R R +S
Sbjct: 263 PYYSRYRSRSRSRS 276
>TR2A_MOUSE (Q6PFR5) Transformer-2 protein homolog (TRA-2 alpha)
Length = 281
Score = 63.9 bits (154), Expect = 4e-10
Identities = 42/144 (29%), Positives = 68/144 (47%), Gaps = 7/144 (4%)
Query: 99 RARVHKLRLEKGAAKAAEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESY 158
R+R + + +++ + H + DP L V LS TTE ++ F Y
Sbjct: 81 RSRSYTPEYRRRRSRSHSPMSNRRRHTGSRANPDPNTCLGVFGLSLYTTERDLREVFSRY 140
Query: 159 GAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVD---VERGRT 215
G + V +V D + + RG+AF+ + D K A ++A+G +++GRR+ VD +R T
Sbjct: 141 GPLSGVNVVYDQRTGRSRGFAFVYFERIDDSKEAMERANGMELDGRRIRVDYSITKRAHT 200
Query: 216 ----VPNWRPRRLGGGLGTTRVGG 235
+ RP GGG G GG
Sbjct: 201 PTPGIYMGRPTHSGGGGGGGGGGG 224
>NUCL_HUMAN (P19338) Nucleolin (Protein C23)
Length = 706
Score = 63.5 bits (153), Expect = 5e-10
Identities = 45/131 (34%), Positives = 61/131 (46%), Gaps = 15/131 (11%)
Query: 115 AEELEKYDPHNDPNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNK 174
A LE P PN P KTLFV LS +TTE +K ES+ R R+VTD E+
Sbjct: 551 AIRLELQGPRGSPNARSQPSKTLFVKGLSEDTTEETLK---ESFDGSVRARIVTDRETGS 607
Query: 175 PRGYAFIEYLHTRDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVG 234
+G+ F+++ D K A + DG +I+G +V +D W + GG G G
Sbjct: 608 SKGFGFVDFNSEEDAKEAME--DG-EIDGNKVTLD---------WAKPKGEGGFGGRGGG 655
Query: 235 GDEVNQRHSGR 245
R GR
Sbjct: 656 RGGFGGRGGGR 666
Score = 56.6 bits (135), Expect = 6e-08
Identities = 32/86 (37%), Positives = 47/86 (54%), Gaps = 4/86 (4%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
KTL ++ LSY TE ++ FE IK V ++ K +GYAFIE+ D K A
Sbjct: 485 KTLVLSNLSYSATEETLQEVFEKATFIK----VPQNQNGKSKGYAFIEFASFEDAKEALN 540
Query: 195 QADGRKIEGRRVLVDVERGRTVPNWR 220
+ R+IEGR + ++++ R PN R
Sbjct: 541 SCNKREIEGRAIRLELQGPRGSPNAR 566
Score = 40.8 bits (94), Expect = 0.004
Identities = 20/72 (27%), Positives = 37/72 (50%), Gaps = 5/72 (6%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
+TL L Y+ T+ +K FE I+ V ++ K +G A+IE+ D + ++
Sbjct: 392 RTLLAKNLPYKVTQDELKEVFEDAAEIRLV-----SKDGKSKGIAYIEFKTEADAEKTFE 446
Query: 195 QADGRKIEGRRV 206
+ G +I+GR +
Sbjct: 447 EKQGTEIDGRSI 458
>SFR2_HUMAN (Q01130) Splicing factor, arginine/serine-rich 2
(Splicing factor SC35) (SC-35) (Splicing component, 35
kDa) (PR264 protein)
Length = 220
Score = 62.8 bits (151), Expect = 9e-10
Identities = 48/157 (30%), Positives = 71/157 (44%), Gaps = 13/157 (8%)
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P++ G +L V L+Y T+ ++R FE Y + V + D + + RG+AF+ +
Sbjct: 7 PDVEG--MTSLKVDNLTYRTSPDTLRRVFEKYRRVGDVYIPRDRYTKESRGFAFVRFHDK 64
Query: 187 RDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWR-------PRRLGGGLGTTRVGGDEVN 239
RD + A DG ++GR + V + R P+ PRR GGG G R
Sbjct: 65 RDAEDAMDAMDGAVLDGRELRVQMARYGRPPDSHHSRRGPPPRRYGGG-GYGRRSRSPRR 123
Query: 240 QRHSGREQQQSGPSRSEEPRVREERHADRDREKSRER 276
+R S + SRS R R R R R +SR R
Sbjct: 124 RRRSRSRSRSRSRSRS---RSRYSRSKSRSRTRSRSR 157
>SFR2_CHICK (P30352) Splicing factor, arginine/serine-rich 2
(Splicing factor SC35) (SC-35) (Splicing component, 35
kDa) (PR264 protein)
Length = 221
Score = 62.4 bits (150), Expect = 1e-09
Identities = 43/153 (28%), Positives = 69/153 (44%), Gaps = 5/153 (3%)
Query: 127 PNISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHT 186
P++ G +L V L+Y T+ ++R FE YG + V + D + + RG+AF+ +
Sbjct: 8 PDVEG--MTSLKVDNLTYRTSPDTLRRVFEKYGRVGDVYIPRDRYTKESRGFAFVRFHDK 65
Query: 187 RDMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLG---GGLGTTRVGGDEVNQRHS 243
RD + A DG ++GR + V + R P+ R G G++ G + R
Sbjct: 66 RDAEDAMDAMDGAVLDGRELRVQMARYGRPPDSHHSRRGPPPRRYGSSGYGRRSRSPRRR 125
Query: 244 GREQQQSGPSRSEEPRVREERHADRDREKSRER 276
R + +S R R R R R +SR R
Sbjct: 126 RRSRSRSRSRSRSRSRSRYSRSKSRSRTRSRSR 158
>NUCL_MOUSE (P09405) Nucleolin (Protein C23)
Length = 706
Score = 62.0 bits (149), Expect = 2e-09
Identities = 40/118 (33%), Positives = 58/118 (48%), Gaps = 12/118 (10%)
Query: 128 NISGDPYKTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTR 187
N P KTLFV LS +TTE +K FE G++ R R+VTD E+ +G+ F+++
Sbjct: 561 NSRSQPSKTLFVKGLSEDTTEETLKESFE--GSV-RARIVTDRETGSSKGFGFVDFNSEE 617
Query: 188 DMKAAYKQADGRKIEGRRVLVDVERGRTVPNWRPRRLGGGLGTTRVGGDEVNQRHSGR 245
D KAA + + +I+G +V +D W + GG G G R GR
Sbjct: 618 DAKAAKEAMEDGEIDGNKVTLD---------WAKPKGEGGFGGRGGGRGGFGGRGGGR 666
Score = 49.3 bits (116), Expect = 1e-05
Identities = 27/77 (35%), Positives = 41/77 (53%), Gaps = 4/77 (5%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
KTL ++ LSY T+ ++ FE IK V KP+GYAFIE+ D K A
Sbjct: 486 KTLVLSNLSYSATKETLEEVFEKATFIK----VPQNPHGKPKGYAFIEFASFEDAKEALN 541
Query: 195 QADGRKIEGRRVLVDVE 211
+ +IEGR + ++++
Sbjct: 542 SCNKMEIEGRTIRLELQ 558
Score = 39.7 bits (91), Expect = 0.008
Identities = 23/72 (31%), Positives = 37/72 (50%), Gaps = 5/72 (6%)
Query: 135 KTLFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYK 194
+TL LS+ TE +K FE +RLV+ + K +G A+IE+ D + +
Sbjct: 394 RTLLAKNLSFNITEDELKEVFED---AMEIRLVS--QDGKSKGIAYIEFKSEADAEKNLE 448
Query: 195 QADGRKIEGRRV 206
+ G +I+GR V
Sbjct: 449 EKQGAEIDGRSV 460
>CIRP_RAT (P60825) Cold-inducible RNA-binding protein (Glycine-rich
RNA-binding protein CIRP) (A18 hnRNP)
Length = 172
Score = 62.0 bits (149), Expect = 2e-09
Identities = 41/129 (31%), Positives = 66/129 (50%), Gaps = 17/129 (13%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV LS++T E +++ F YG I V +V D E+ + RG+ F+ + + D K A
Sbjct: 8 LFVGGLSFDTNEQALEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAM 67
Query: 197 DGRKIEGRRVLVDVERGRTVPN----WRPRRLGG----------GLGTTRVGGDEVNQRH 242
+G+ ++GR++ VD + G++ N +R GG G G +R GGD
Sbjct: 68 NGKSVDGRQIRVD-QAGKSSDNRSRGYRGGSAGGRGFFRGGRSRGRGFSRGGGD--RGYG 124
Query: 243 SGREQQQSG 251
GR + +SG
Sbjct: 125 GGRFESRSG 133
>CIRP_MOUSE (P60824) Cold-inducible RNA-binding protein
(Glycine-rich RNA-binding protein CIRP) (A18 hnRNP)
Length = 172
Score = 62.0 bits (149), Expect = 2e-09
Identities = 41/129 (31%), Positives = 66/129 (50%), Gaps = 17/129 (13%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV LS++T E +++ F YG I V +V D E+ + RG+ F+ + + D K A
Sbjct: 8 LFVGGLSFDTNEQALEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAM 67
Query: 197 DGRKIEGRRVLVDVERGRTVPN----WRPRRLGG----------GLGTTRVGGDEVNQRH 242
+G+ ++GR++ VD + G++ N +R GG G G +R GGD
Sbjct: 68 NGKSVDGRQIRVD-QAGKSSDNRSRGYRGGSAGGRGFFRGGRSRGRGFSRGGGD--RGYG 124
Query: 243 SGREQQQSG 251
GR + +SG
Sbjct: 125 GGRFESRSG 133
>CIRP_CRIGR (P60826) Cold-inducible RNA-binding protein
(Glycine-rich RNA-binding protein CIRP) (A18 hnRNP)
Length = 172
Score = 62.0 bits (149), Expect = 2e-09
Identities = 41/129 (31%), Positives = 66/129 (50%), Gaps = 17/129 (13%)
Query: 137 LFVARLSYETTESRIKREFESYGAIKRVRLVTDAESNKPRGYAFIEYLHTRDMKAAYKQA 196
LFV LS++T E +++ F YG I V +V D E+ + RG+ F+ + + D K A
Sbjct: 8 LFVGGLSFDTNEQALEQVFSKYGQISEVVVVKDRETQRSRGFGFVTFENIDDAKDAMMAM 67
Query: 197 DGRKIEGRRVLVDVERGRTVPN----WRPRRLGG----------GLGTTRVGGDEVNQRH 242
+G+ ++GR++ VD + G++ N +R GG G G +R GGD
Sbjct: 68 NGKSVDGRQIRVD-QAGKSSDNRSRGYRGGSAGGRGFFRGGRSRGRGFSRGGGD--RGYG 124
Query: 243 SGREQQQSG 251
GR + +SG
Sbjct: 125 GGRFESRSG 133
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 33,969,700
Number of Sequences: 164201
Number of extensions: 1538467
Number of successful extensions: 5600
Number of sequences better than 10.0: 364
Number of HSP's better than 10.0 without gapping: 256
Number of HSP's successfully gapped in prelim test: 108
Number of HSP's that attempted gapping in prelim test: 4668
Number of HSP's gapped (non-prelim): 797
length of query: 276
length of database: 59,974,054
effective HSP length: 109
effective length of query: 167
effective length of database: 42,076,145
effective search space: 7026716215
effective search space used: 7026716215
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147964.4


