

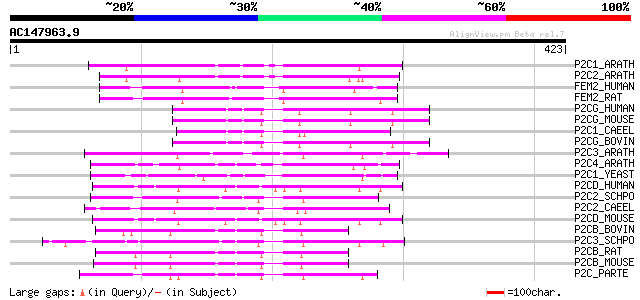
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147963.9 - phase: 0
(423 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
P2C1_ARATH (P49597) Protein phosphatase 2C ABI1 (EC 3.1.3.16) (P... 96 2e-19
P2C2_ARATH (O04719) Protein phosphatase 2C ABI2 (EC 3.1.3.16) (P... 91 4e-18
FEM2_HUMAN (P49593) Ca(2+)/calmodulin-dependent protein kinase p... 83 2e-15
FEM2_RAT (Q9WVR7) Ca(2+)/calmodulin-dependent protein kinase pho... 77 6e-14
P2CG_HUMAN (O15355) Protein phosphatase 2C gamma isoform (EC 3.1... 75 2e-13
P2CG_MOUSE (Q61074) Protein phosphatase 2C gamma isoform (EC 3.1... 74 7e-13
P2C1_CAEEL (P49595) Probable protein phosphatase 2C F42G9.1 (EC ... 74 9e-13
P2CG_BOVIN (P79126) Protein phosphatase 2C gamma isoform (EC 3.1... 73 1e-12
P2C3_ARATH (P49599) Protein phosphatase 2C PPH1 (EC 3.1.3.16) (P... 72 2e-12
P2C4_ARATH (P49598) Protein phosphatase 2C (EC 3.1.3.16) (PP2C) 70 1e-11
P2C1_YEAST (P35182) Protein phosphatase 2C homolog 1 (EC 3.1.3.1... 70 1e-11
P2CD_HUMAN (O15297) Protein phosphatase 2C delta isoform (EC 3.1... 69 2e-11
P2C2_SCHPO (Q09172) Protein phosphatase 2C homolog 2 (EC 3.1.3.1... 69 3e-11
P2C2_CAEEL (P49596) Probable protein phosphatase 2C T23F11.1 (EC... 68 4e-11
P2CD_MOUSE (Q9QZ67) Protein phosphatase 2C delta isoform (EC 3.1... 68 5e-11
P2CB_BOVIN (O62830) Protein phosphatase 2C beta isoform (EC 3.1.... 68 5e-11
P2C3_SCHPO (Q09173) Protein phosphatase 2C homolog 3 (EC 3.1.3.1... 68 5e-11
P2CB_RAT (P35815) Protein phosphatase 2C beta isoform (EC 3.1.3.... 67 9e-11
P2CB_MOUSE (P36993) Protein phosphatase 2C beta isoform (EC 3.1.... 67 9e-11
P2C_PARTE (P49444) Protein phosphatase 2C (EC 3.1.3.16) (PP2C) 67 1e-10
>P2C1_ARATH (P49597) Protein phosphatase 2C ABI1 (EC 3.1.3.16)
(PP2C) (Abscisic acid-insensitive 1)
Length = 434
Score = 95.9 bits (237), Expect = 2e-19
Identities = 76/271 (28%), Positives = 123/271 (45%), Gaps = 43/271 (15%)
Query: 61 DSSTSFSVFAILDGHNGISAAIFAKEN----IINNVMSAIPQGVSREEWLQALPRALVVA 116
D ++ F + DGH G A + +E + + P + WL+ +AL +
Sbjct: 165 DPQSAAHFFGVYDGHGGSQVANYCRERMHLALAEEIAKEKPMLCDGDTWLEKWKKALFNS 224
Query: 117 FVKTDMEFQKKG-ETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEE 175
F++ D E + ET G+T+ ++ + VA+ GDSR +L +G L+VDH+ +
Sbjct: 225 FLRVDSEIESVAPETVGSTSVVAVVFPSHIFVANCGDSRAVL-CRGKTALPLSVDHKPDR 283
Query: 176 NVEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQV 235
+E R+ A+GG+V + N G V G L +SRSIGD + I+P P V V
Sbjct: 284 E-DEAARIEAAGGKVIQWN---GARVF------GVLAMSRSIGDRYLKPSIIPDPEVTAV 333
Query: 236 KLSNAGGRLIIASDGIWDTLSSDMAAKSCR---------------------------GVP 268
K LI+ASDG+WD ++ + A + R P
Sbjct: 334 KRVKEDDCLILASDGVWDVMTDEEACEMARKRILLWHKKNAVAGDASLLADERRKEGKDP 393
Query: 269 AELAAKLVVKEALRSRGLKDDTTCLVVDIIP 299
A ++A + + RG KD+ + +VVD+ P
Sbjct: 394 AAMSAAEYLSKLAIQRGSKDNISVVVVDLKP 424
>P2C2_ARATH (O04719) Protein phosphatase 2C ABI2 (EC 3.1.3.16)
(PP2C) (Abscisic acid-insensitive 2)
Length = 423
Score = 91.3 bits (225), Expect = 4e-18
Identities = 74/262 (28%), Positives = 124/262 (47%), Gaps = 44/262 (16%)
Query: 69 FAILDGHNGISAAIFAKEN----IINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEF 124
F + DGH G A + +E + ++ P+ + W + +AL +F++ D E
Sbjct: 161 FGVYDGHGGSQVANYCRERMHLALTEEIVKEKPEFCDGDTWQEKWKKALFNSFMRVDSEI 220
Query: 125 QKKG---ETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERE 181
+ ET G+T+ ++ + VA+ GDSR +L +G L+VDH+ + + +E
Sbjct: 221 ETVAHAPETVGSTSVVAVVFPTHIFVANCGDSRAVL-CRGKTPLALSVDHKPDRD-DEAA 278
Query: 182 RVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAG 241
R+ A+GG+V R N G V G L +SRSIGD + ++P P V V+
Sbjct: 279 RIEAAGGKVIRWN---GARVF------GVLAMSRSIGDRYLKPSVIPDPEVTSVRRVKED 329
Query: 242 GRLIIASDGIWDTLSS----DMAAKSC-------------------RGV---PAELAAKL 275
LI+ASDG+WD +++ D+A K RG PA ++A
Sbjct: 330 DCLILASDGLWDVMTNEEVCDLARKRILLWHKKNAMAGEALLPAEKRGEGKDPAAMSAAE 389
Query: 276 VVKEALRSRGLKDDTTCLVVDI 297
+ + +G KD+ + +VVD+
Sbjct: 390 YLSKMALQKGSKDNISVVVVDL 411
>FEM2_HUMAN (P49593) Ca(2+)/calmodulin-dependent protein kinase
phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase)
(CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein
phosphatase 1F)
Length = 454
Score = 82.8 bits (203), Expect = 2e-15
Identities = 77/241 (31%), Positives = 111/241 (45%), Gaps = 37/241 (15%)
Query: 69 FAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPR-ALVVAFVKTDMEFQKK 127
FA+ DGH G+ AA +A ++ N +R+ L P AL AF +TD F +K
Sbjct: 194 FAVFDGHGGVDAARYAAVHVHTNA--------ARQPELPTDPEGALREAFRRTDQMFLRK 245
Query: 128 GET----SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERV 183
+ SGTT +I G T+ VA +GDS+ IL QG VV L+ HR E +E+ R+
Sbjct: 246 AKRERLQSGTTGVCALIAGATLHVAWLGDSQVILVQQGQVVKLME-PHR-PERQDEKARI 303
Query: 184 TASGGEVGRLNVFGGNEVGPLRCW--PGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAG 241
A GG V ++ CW G L +SR+IGD Y+ L+ +
Sbjct: 304 EALGGFVSHMD-----------CWRVNGTLAVSRAIGDVFQKPYVSGEADAASRALTGSE 352
Query: 242 GRLIIASDGIWDTLSSDMAA-------KSCRGVPAELAAKLVVKEALRSRGLKDDTTCLV 294
L++A DG +D + +G +A +LV A R RG D+ T +V
Sbjct: 353 DYLLLACDGFFDVVPHQEVVGLVQSHLTRQQGSGLRVAEELVA--AARERGSHDNITVMV 410
Query: 295 V 295
V
Sbjct: 411 V 411
>FEM2_RAT (Q9WVR7) Ca(2+)/calmodulin-dependent protein kinase
phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase)
(CaMKPase) (Partner of PIX 2) (Protein phosphatase 1F)
Length = 450
Score = 77.4 bits (189), Expect = 6e-14
Identities = 72/238 (30%), Positives = 106/238 (44%), Gaps = 31/238 (13%)
Query: 69 FAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKG 128
FA+ DGH G+ AA +A ++ N + E L AL AF TD F +K
Sbjct: 190 FAVFDGHGGVDAARYASVHVHTNASH-------QPELLTDPAAALKEAFRHTDQMFLQKA 242
Query: 129 ET----SGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVT 184
+ SGTT +I G + VA +GDS+ IL QG VV L+ + E +E+ R+
Sbjct: 243 KRERLQSGTTGVCALITGAALHVAWLGDSQVILVQQGQVVKLM--EPHKPERQDEKSRIE 300
Query: 185 ASGGEVGRLNVFGGNEVGPLRCW--PGGLCLSRSIGDTDVGEYIVPIPHVKQVKLSNAGG 242
A GG V ++ CW G L +SR+IGD Y+ +L+
Sbjct: 301 ALGGFVSLMD-----------CWRVNGTLAVSRAIGDVFQKPYVSGEADAASRELTGLED 349
Query: 243 RLIIASDGIWDTLSSDMAAKSCRG-VPAELAAKLVVKEAL----RSRGLKDDTTCLVV 295
L++A DG +D + G + + + + V E L R RG D+ T +VV
Sbjct: 350 YLLLACDGFFDVVPHHEIPGLVHGHLLRQKGSGMHVAEELVAVARDRGSHDNITVMVV 407
>P2CG_HUMAN (O15355) Protein phosphatase 2C gamma isoform (EC
3.1.3.16) (PP2C-gamma) (Protein phosphatase
magnesium-dependent 1 gamma) (Protein phosphatase 1C)
Length = 546
Score = 75.5 bits (184), Expect = 2e-13
Identities = 67/224 (29%), Positives = 103/224 (45%), Gaps = 44/224 (19%)
Query: 125 QKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVT 184
++ G SGTTA +I G + VA+ GDSRC++ G + ++ DH+ E+ V E R+
Sbjct: 321 EEPGSDSGTTAVVALIRGKQLIVANAGDSRCVVSEAGKALD-MSYDHKPEDEV-ELARIK 378
Query: 185 ASGGEV---GRLNVFGGNEVGPLRCWPGGLCLSRSIGD---------TDVGEYIVPIPHV 232
+GG+V GR+N GGL LSR+IGD + I +P +
Sbjct: 379 NAGGKVTMDGRVN--------------GGLNLSRAIGDHFYKRNKNLPPEEQMISALPDI 424
Query: 233 KQVKLSNAGGRLIIASDGIWDTLSSD------MAAKSCRGVPAELAAKLVVKEALRSRGL 286
K + L++ ++IA DGIW+ +SS + S R EL + E L + L
Sbjct: 425 KVLTLTDDHEFMVIACDGIWNVMSSQEVVDFIQSKISQRDENGELRLLSSIVEELLDQCL 484
Query: 287 KDDT----------TCLVVDIIPSDYPVLPMPATPRKKHNVLSS 320
DT TC+++ P + L + RK VLS+
Sbjct: 485 APDTSGDGTGCDNMTCIIICFKPRNTAELQPESGKRKLEEVLST 528
>P2CG_MOUSE (Q61074) Protein phosphatase 2C gamma isoform (EC
3.1.3.16) (PP2C-gamma) (Protein phosphatase
magnesium-dependent 1 gamma) (Protein phosphatase 1C)
(Fibroblast growth factor inducible protein 13) (FIN13)
Length = 542
Score = 73.9 bits (180), Expect = 7e-13
Identities = 66/224 (29%), Positives = 102/224 (45%), Gaps = 44/224 (19%)
Query: 125 QKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVT 184
++ G SGTTA +I G + VA+ GDSRC++ G + ++ DH+ E+ V E R+
Sbjct: 318 EEPGSDSGTTAVVALIRGKQLIVANAGDSRCVVSEAGKALD-MSYDHKPEDEV-ELARIK 375
Query: 185 ASGGEV---GRLNVFGGNEVGPLRCWPGGLCLSRSIGD---------TDVGEYIVPIPHV 232
+GG+V GR+N GGL LSR+IGD + I +P +
Sbjct: 376 NAGGKVTMDGRVN--------------GGLNLSRAIGDHFYKRNKNLPPQEQMISALPDI 421
Query: 233 KQVKLSNAGGRLIIASDGIWDTLSSD------MAAKSCRGVPAELAAKLVVKEALRSRGL 286
K + L++ ++IA DGIW+ +SS + S R EL + E L + L
Sbjct: 422 KVLTLTDDHEFMVIACDGIWNVMSSQEVVDFIQSKISQRDENGELRLLSSIVEELLDQCL 481
Query: 287 KDDT----------TCLVVDIIPSDYPVLPMPATPRKKHNVLSS 320
DT TC+++ P + L + RK LS+
Sbjct: 482 APDTSGDGTGCDNMTCIIICFKPRNTVELQAESGKRKLEEALST 525
>P2C1_CAEEL (P49595) Probable protein phosphatase 2C F42G9.1 (EC
3.1.3.16) (PP2C)
Length = 491
Score = 73.6 bits (179), Expect = 9e-13
Identities = 59/174 (33%), Positives = 88/174 (49%), Gaps = 27/174 (15%)
Query: 128 GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASG 187
GE SGTTA ++ V VA+ GDSR +L G V L+VDH+ E+ V E R+ A+G
Sbjct: 312 GEDSGTTACVCLVGKDKVIVANAGDSRAVLCRNGKAVD-LSVDHKPEDEV-ETNRIHAAG 369
Query: 188 GEV--GRLNVFGGNEVGPLRCWPGGLCLSRSIGD------TDVG---EYIVPIPHVKQVK 236
G++ GR+N GGL LSR+ GD ++G + I +P VK
Sbjct: 370 GQIEDGRVN--------------GGLNLSRAFGDHAYKKNQELGLKEQMITALPDVKIEA 415
Query: 237 LSNAGGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEALRSRGLKDDT 290
L+ +++A DGIW+++ S R + A+ ++ V +AL L D T
Sbjct: 416 LTPEDEFIVVACDGIWNSMESQQVVDFVRDLLAKGSSCAEVCDALCDACLADST 469
>P2CG_BOVIN (P79126) Protein phosphatase 2C gamma isoform (EC
3.1.3.16) (PP2C-gamma) (Protein phosphatase
magnesium-dependent 1 gamma) (Protein phosphatase 1B)
(Magnesium-dependent calcium inhibitable phosphatase)
(MCPP)
Length = 543
Score = 73.2 bits (178), Expect = 1e-12
Identities = 66/224 (29%), Positives = 102/224 (45%), Gaps = 44/224 (19%)
Query: 125 QKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVT 184
++ G SGTTA +I G + VA+ GDSRC++ G + ++ DH+ E+ V E R+
Sbjct: 319 EEPGSDSGTTAVVALIRGKQLIVANAGDSRCVVSEAGKALD-MSYDHKPEDEV-ELARIK 376
Query: 185 ASGGEV---GRLNVFGGNEVGPLRCWPGGLCLSRSIGD---------TDVGEYIVPIPHV 232
+GG+V GR+N GGL LSR+IGD + I +P +
Sbjct: 377 NAGGKVTMDGRVN--------------GGLNLSRAIGDHFYKRNKNLPPEEQMISALPDI 422
Query: 233 KQVKLSNAGGRLIIASDGIWDTLSSD------MAAKSCRGVPAELAAKLVVKEALRSRGL 286
K + L++ ++IA DGIW+ +SS + S R EL + E L + L
Sbjct: 423 KVLTLTDDHEFMVIACDGIWNVMSSQEVIDFIQSKISQRDENGELRLLSSIVEYLLDQCL 482
Query: 287 KDDT----------TCLVVDIIPSDYPVLPMPATPRKKHNVLSS 320
DT TC+++ P + + RK VLS+
Sbjct: 483 APDTSGDGTGCDNMTCIIICFKPRNTAAPQPESGKRKLEEVLST 526
>P2C3_ARATH (P49599) Protein phosphatase 2C PPH1 (EC 3.1.3.16)
(PP2C)
Length = 388
Score = 72.4 bits (176), Expect = 2e-12
Identities = 80/318 (25%), Positives = 134/318 (41%), Gaps = 56/318 (17%)
Query: 58 VPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQG-VSREEWLQALPRALVVA 116
+ D+ SFS A+ DGH G S+ F +E + + A+ G + A+ AL+ A
Sbjct: 78 IRSDAVDSFSYAAVFDGHAGSSSVKFLREELYKECVGALQAGSLLNGGDFAAIKEALIKA 137
Query: 117 FVKTDMEFQK-------KGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTV 169
F D K + + SG+TAT +II +A +GDS C + ++ G + LT
Sbjct: 138 FESVDRNLLKWLEANGDEEDESGSTATVMIIRNDVSFIAHIGDS-CAVLSRSGQIEELTD 196
Query: 170 DHRLEENVEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDV------- 222
HR + A+ EV R+ GG V C G + +SR+ GD
Sbjct: 197 YHRPYGSSR------AAIQEVKRVKEAGGWIVNGRIC--GDIAVSRAFGDIRFKTKKNDM 248
Query: 223 ---------------------GEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAA 261
G+ +V P + QV L++ +I+ASDG+WD + S
Sbjct: 249 LKKGVDEGRWSEKFVSRIEFKGDMVVATPDIFQVPLTSDVEFIILASDGLWDYMKSSDVV 308
Query: 262 KSCRGV-----PAELAAKLVVKEALRSRGLKDDTTCLVVDIIPSDYPVLPMPATPRKKHN 316
R +LA + + + AL R +D+ + ++ D+ +++ L P ++ N
Sbjct: 309 SYVRDQLRKHGNVQLACESLAQVALDRRS-QDNISIIIADLGRTEWKNL-----PAQRQN 362
Query: 317 VLSSLLFGKKSQNLANKG 334
V+ L+ + L G
Sbjct: 363 VVVELVQAATTIGLVTVG 380
>P2C4_ARATH (P49598) Protein phosphatase 2C (EC 3.1.3.16) (PP2C)
Length = 399
Score = 69.7 bits (169), Expect = 1e-11
Identities = 72/276 (26%), Positives = 125/276 (45%), Gaps = 57/276 (20%)
Query: 62 SSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTD 121
+S + + + DGH A +E + + V + + ++ +EW + + V +F K D
Sbjct: 131 NSENHHFYGVFDGHGCSHVAEKCRERLHDIVKKEV-EVMASDEWTETM----VKSFQKMD 185
Query: 122 MEFQKKG-------------------------ETSGTTATFVIIDGWTVTVASVGDSRCI 156
E ++ + G+TA ++ + V++ GDSR +
Sbjct: 186 KEVSQRECNLVVNGATRSMKNSCRCELQSPQCDAVGSTAVVSVVTPEKIIVSNCGDSRAV 245
Query: 157 LDTQGGVVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRS 216
L + GV L+VDH+ + +E R+ +GG R+ + G V G L +SR+
Sbjct: 246 L-CRNGVAIPLSVDHK-PDRPDELIRIQQAGG---RVIYWDGARV------LGVLAMSRA 294
Query: 217 IGDTDVGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMA---AKSC-RGVPA--- 269
IGD + Y++P P V ++ LI+ASDG+WD + ++ A A+ C RG A
Sbjct: 295 IGDNYLKPYVIPDPEVTVTDRTDEDECLILASDGLWDVVPNETACGVARMCLRGAGAGDD 354
Query: 270 --------ELAAKLVVKEALRSRGLKDDTTCLVVDI 297
AA L+ K AL +R D+ + +VVD+
Sbjct: 355 SDAAHNACSDAALLLTKLAL-ARQSSDNVSVVVVDL 389
>P2C1_YEAST (P35182) Protein phosphatase 2C homolog 1 (EC 3.1.3.16)
(PP2C-1)
Length = 281
Score = 69.7 bits (169), Expect = 1e-11
Identities = 70/255 (27%), Positives = 117/255 (45%), Gaps = 43/255 (16%)
Query: 62 SSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTD 121
S + FA+ DGH GI A+ + +++ + I Q + +E + + L +F+ D
Sbjct: 47 SRLDWGYFAVFDGHAGIQASKWCGKHL----HTIIEQNILADE-TRDVRDVLNDSFLAID 101
Query: 122 MEFQKK-GETSGTTATFVIIDGWTVT------------------VASVGDSRCILDTQGG 162
E K SG TA ++ W + A+VGDSR +L G
Sbjct: 102 EEINTKLVGNSGCTAAVCVLR-WELPDSVSDDSMDLAQHQRKLYTANVGDSRIVLFRNGN 160
Query: 163 VVSLLTVDHRLEENVEERERVTASGGEVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDV 222
+ L T DH+ + +E +RV +GG + + V G L ++RS+GD
Sbjct: 161 SIRL-TYDHKASDTLE-MQRVEQAGGLIMKSRV------------NGMLAVTRSLGDKFF 206
Query: 223 GEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGV--PAELAAKLVVKEA 280
+V P V++++ LI+A DG+WD + A + + + P E AAK++V+ A
Sbjct: 207 DSLVVGSPFTTSVEITSEDKFLILACDGLWDVIDDQDACELIKDITEPNE-AAKVLVRYA 265
Query: 281 LRSRGLKDDTTCLVV 295
L + G D+ T +VV
Sbjct: 266 LEN-GTTDNVTVMVV 279
>P2CD_HUMAN (O15297) Protein phosphatase 2C delta isoform (EC
3.1.3.16) (PP2C-delta) (p53-induced protein phosphatase
1) (Protein phosphatase magnesium-dependent 1 delta)
Length = 605
Score = 69.3 bits (168), Expect = 2e-11
Identities = 85/288 (29%), Positives = 128/288 (43%), Gaps = 58/288 (20%)
Query: 64 TSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDME 123
+S + FA+ DGH G AA FA+E++ + +G + E + A+ F+ +
Sbjct: 96 SSVAFFAVCDGHGGREAAQFAREHLWGFIKKQ--KGFTSSEPAKVCA-AIRKGFLACHLA 152
Query: 124 FQKK-----------GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGV------VSL 166
KK TSGTTA+ VII G + VA VGDS +L Q
Sbjct: 153 MWKKLAEWPKTMTGLPSTSGTTASVVIIRGMKMYVAHVGDSGVVLGIQDDPKDDFVRAVE 212
Query: 167 LTVDHRLEENVEERERVTASGGEVGRLNVFGGNEV----------GPLRCWP-----GGL 211
+T DH+ E +ERER+ GG V +N G N V GP+R L
Sbjct: 213 VTQDHK-PELPKERERIEGLGGSV--MNKSGVNRVVWKRPRLTHNGPVRRSTVIDQIPFL 269
Query: 212 CLSRSIGDT-----DVGEYIV-PIPHVKQVKLSNAGGR-LIIASDGIWDTLSSDMAAKSC 264
++R++GD GE++V P P L + +I+ SDG+W+ + A C
Sbjct: 270 AVARALGDLWSYDFFSGEFVVSPEPDTSVHTLDPQKHKYIILGSDGLWNMIPPQDAISMC 329
Query: 265 R---------GVPAELAAKLVVKEAL---RSRGLK-DDTTCLVVDIIP 299
+ G + AK++V AL R R L+ D+T+ +V+ I P
Sbjct: 330 QDQEEKKYLMGEHGQSCAKMLVNRALGRWRQRMLRADNTSAIVICISP 377
>P2C2_SCHPO (Q09172) Protein phosphatase 2C homolog 2 (EC 3.1.3.16)
(PP2C-2)
Length = 370
Score = 68.6 bits (166), Expect = 3e-11
Identities = 64/237 (27%), Positives = 105/237 (44%), Gaps = 39/237 (16%)
Query: 62 SSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTD 121
S+ S F + DGH G A + ++++ + + S W AL F+ D
Sbjct: 52 SNPPTSFFGVFDGHGGDRVAKYCRQHLPDIIKS------QPSFWKGNYDEALKSGFLAAD 105
Query: 122 MEFQK----KGETSGTTATF-VIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEEN 176
+ + + SG TAT +I+D + A+ GDSR +L + G L+ DH+ +
Sbjct: 106 NALMQDRDMQEDPSGCTATTALIVDHQVIYCANAGDSRTVLGRK-GTAEPLSFDHKPNND 164
Query: 177 VEERERVTASGG--EVGRLNVFGGNEVGPLRCWPGGLCLSRSIGDTDV---------GEY 225
V E+ R+TA+GG + GR+N G L LSR+IGD + +
Sbjct: 165 V-EKARITAAGGFIDFGRVN--------------GSLALSRAIGDFEYKKDSSLPPEKQI 209
Query: 226 IVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSC-RGVPAELAAKLVVKEAL 281
+ P V + LI+A DGIWD SS + RG+ A + +++ + +
Sbjct: 210 VTAFPDVVIHNIDPDDEFLILACDGIWDCKSSQQVVEFVRRGIVARQSLEVICENLM 266
>P2C2_CAEEL (P49596) Probable protein phosphatase 2C T23F11.1 (EC
3.1.3.16) (PP2C)
Length = 356
Score = 68.2 bits (165), Expect = 4e-11
Identities = 69/248 (27%), Positives = 109/248 (43%), Gaps = 41/248 (16%)
Query: 58 VPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQA-LPRALVVA 116
+P D +F FA+ DGH G + ++ N+ V V+++E+ + + A+
Sbjct: 46 LPDDPKCAF--FAVYDGHGGSKVSQYSGINLHKKV-------VAQKEFSEGNMKEAIEKG 96
Query: 117 FVKTDMEF----QKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHR 172
F++ D + + K + SGTTA V+I V + GDSR + + G L+ DH+
Sbjct: 97 FLELDQQMRVDEETKDDVSGTTAVVVLIKEGDVYCGNAGDSRAV-SSVVGEARPLSFDHK 155
Query: 173 LEENVEERERVTASGG--EVGRLNVFGGNEVGPLRCWPGGLCLSRSIG-------DTDVG 223
E R R+ A+GG E R+N G L LSR++G DT
Sbjct: 156 PSHETEAR-RIIAAGGWVEFNRVN--------------GNLALSRALGDFAFKNCDTKPA 200
Query: 224 E--YIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGVPAELAAKLVVKEAL 281
E + P V KL+ +++A DGIWD +++ R AE + E L
Sbjct: 201 EEQIVTAFPDVITDKLTPDHEFIVLACDGIWDVMTNQEVVDFVREKLAEKRDPQSICEEL 260
Query: 282 RSRGLKDD 289
+R L D
Sbjct: 261 LTRCLAPD 268
>P2CD_MOUSE (Q9QZ67) Protein phosphatase 2C delta isoform (EC
3.1.3.16) (PP2C-delta) (p53-induced protein phosphatase
1) (Protein phosphatase magnesium-dependent 1 delta)
Length = 598
Score = 67.8 bits (164), Expect = 5e-11
Identities = 84/288 (29%), Positives = 128/288 (44%), Gaps = 58/288 (20%)
Query: 64 TSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDME 123
+S + FA+ DGH G AA FA+E++ + +G + E + A+ F+ +
Sbjct: 89 SSVAFFAVCDGHGGREAAQFAREHLWGFIKKQ--KGFTSSEPAKVCA-AIRKGFLACHLA 145
Query: 124 FQKK-----------GETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGV------VSL 166
KK TSGTTA+ VII G + VA VGDS +L Q
Sbjct: 146 MWKKLAEWPKTMTGLPSTSGTTASVVIIRGMKMYVAHVGDSGVVLGIQDDPKDDFVRAVE 205
Query: 167 LTVDHRLEENVEERERVTASGGEVGRLNVFGGNEV----------GPLRCWP-----GGL 211
+T DH+ E +ERER+ GG V +N G N V GP+R L
Sbjct: 206 VTQDHK-PELPKERERIEGLGGSV--MNKSGVNRVVWKRPRLTHSGPVRRSTVIDQIPFL 262
Query: 212 CLSRSIGDT-----DVGEYIV-PIPHVKQVKLSNAGGR-LIIASDGIWDTLSSDMAAKSC 264
++R++GD G+++V P P L + +I+ SDG+W+ + A C
Sbjct: 263 AVARALGDLWSYDFFSGKFVVSPEPDTSVHTLDPRKHKYIILGSDGLWNMVPPQDAISMC 322
Query: 265 R---------GVPAELAAKLVVKEAL---RSRGLK-DDTTCLVVDIIP 299
+ G + AK++V AL R R L+ D+T+ +V+ I P
Sbjct: 323 QDQEEKKYLMGEQGQSCAKMLVNRALGRWRQRMLRADNTSAIVICISP 370
>P2CB_BOVIN (O62830) Protein phosphatase 2C beta isoform (EC
3.1.3.16) (PP2C-beta)
Length = 387
Score = 67.8 bits (164), Expect = 5e-11
Identities = 61/216 (28%), Positives = 99/216 (45%), Gaps = 39/216 (18%)
Query: 66 FSVFAILDGHNGISAAIFAK----ENIINN--VMSAIPQGVSREEWLQALPRALVVAFVK 119
+S FA+ DGH G A + E+I NN +A G + E ++ + + F+K
Sbjct: 53 WSFFAVYDGHAGSRVANYCSTHLLEHITNNEDFRAAGKSGSALEPSVENVKNGIRTGFLK 112
Query: 120 TD------MEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRL 173
D + + + SG+TA V+I + + GDSR +L + G V T DH+
Sbjct: 113 IDEYMRNFSDLRNGMDRSGSTAVGVMISPKHIYFINCGDSRAVL-YRSGQVCFSTQDHK- 170
Query: 174 EENVEERERVTASGGEV--GRLNVFGGNEVGPLRCWPGGLCLSRSIGDTD---------V 222
N E+ER+ +GG V R+N G L +SR++GD D
Sbjct: 171 PCNPREKERIQNAGGSVMIQRVN--------------GSLAVSRALGDYDYKCVDGKGPT 216
Query: 223 GEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSD 258
+ + P P V ++ + +I+A DGIWD +S++
Sbjct: 217 EQLVSPEPEVYEILRAEEDEFIILACDGIWDVMSNE 252
>P2C3_SCHPO (Q09173) Protein phosphatase 2C homolog 3 (EC 3.1.3.16)
(PP2C-3)
Length = 414
Score = 67.8 bits (164), Expect = 5e-11
Identities = 75/302 (24%), Positives = 131/302 (42%), Gaps = 50/302 (16%)
Query: 26 NGKTEKPFVKYGQAGL-----AKKGEDYFLIKTDCHRVPGDSSTSFSVFAILDGHNGISA 80
NG E FV YG + + + + ++ +C V FA+ DGH G
Sbjct: 16 NGSNE--FVLYGLSSMQGWRISMEDAHSAILSMECSAV----KDPVDFFAVYDGHGGDKV 69
Query: 81 AIFAKENIINNVMSAIPQGVSREEWLQALPRALVVAFVKTDMEFQKKGETSGTTATFVII 140
A + N+ ++ P + +++ AL + + A + Q + SG TAT V+
Sbjct: 70 AKWCGSNL-PQILEKNPD-FQKGDFVNALKSSFLNADKAILDDDQFHTDPSGCTATVVLR 127
Query: 141 DGWTVTVASVGDSRCILDTQGGVVSLLTVDHRLEENVEERERVTASGG--EVGRLNVFGG 198
G + A+ GDSR +L ++ G+ L+ DH+ N E+ R+ A+GG + GR+N
Sbjct: 128 VGNKLYCANAGDSRTVLGSK-GIAKPLSADHK-PSNEAEKARICAAGGFVDFGRVN---- 181
Query: 199 NEVGPLRCWPGGLCLSRSIGDTDV--------GEYIVPIPHVKQVKLSNAGGRLIIASDG 250
G L LSR+IGD + + + +P V ++++ +++A DG
Sbjct: 182 ----------GNLALSRAIGDFEFKNSNLEPEKQIVTALPDVVVHEITDDDEFVVLACDG 231
Query: 251 IWDTLSSDMAAKSCR-----GVPAELAAKLVVKEALRS------RGLKDDTTCLVVDIIP 299
IWD +S + R G E A+ ++ + S G + T C+V +
Sbjct: 232 IWDCKTSQQVIEFVRRGIVAGTSLEKIAENLMDNCIASDTETTGLGCDNMTVCIVALLQE 291
Query: 300 SD 301
+D
Sbjct: 292 ND 293
>P2CB_RAT (P35815) Protein phosphatase 2C beta isoform (EC 3.1.3.16)
(PP2C-beta) (IA) (Protein phosphatase 1B)
Length = 390
Score = 67.0 bits (162), Expect = 9e-11
Identities = 56/216 (25%), Positives = 99/216 (44%), Gaps = 39/216 (18%)
Query: 66 FSVFAILDGHNGISAAIFAKENIINNVMS------AIPQGVSREEWLQALPRALVVAFVK 119
+S FA+ DGH G A + +++ ++ + A G + E ++ + + F+K
Sbjct: 53 WSFFAVYDGHAGSRVANYCSTHLLEHITTNEDFRAADKSGFALEPSVENVKTGIRTGFLK 112
Query: 120 TD------MEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHRL 173
D + + + SG+TA V+I + + GDSR +L + G V T DH+
Sbjct: 113 IDEYMRNFSDLRNGMDRSGSTAVGVMISPTHIYFINCGDSRAVL-CRNGQVCFSTQDHK- 170
Query: 174 EENVEERERVTASGGEV--GRLNVFGGNEVGPLRCWPGGLCLSRSIGDTD---------V 222
N E+ER+ +GG V R+N G L +SR++GD D
Sbjct: 171 PCNPMEKERIQNAGGSVMIQRVN--------------GSLAVSRALGDYDYKCVDGKGPT 216
Query: 223 GEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSD 258
+ + P P V ++ + +++A DGIWD +S++
Sbjct: 217 EQLVSPEPEVYEILRAEEDEFVVLACDGIWDVMSNE 252
>P2CB_MOUSE (P36993) Protein phosphatase 2C beta isoform (EC
3.1.3.16) (PP2C-beta) (IA) (Protein phosphatase 1B)
Length = 390
Score = 67.0 bits (162), Expect = 9e-11
Identities = 55/217 (25%), Positives = 101/217 (46%), Gaps = 39/217 (17%)
Query: 65 SFSVFAILDGHNGISAAIFAKENIINNVMS------AIPQGVSREEWLQALPRALVVAFV 118
++S FA+ DGH G A + +++ ++ + A G + E ++++ + F+
Sbjct: 52 NWSFFAVYDGHAGSRVANYCSTHLLEHITTNEDFRAADKSGSALEPSVESVKTGIRTGFL 111
Query: 119 KTD------MEFQKKGETSGTTATFVIIDGWTVTVASVGDSRCILDTQGGVVSLLTVDHR 172
K D + + + SG+TA V++ + + GDSR +L + G V T DH+
Sbjct: 112 KIDEYMRNFSDLRNGMDRSGSTAVGVMVSPTHMYFINCGDSRAVL-CRNGQVCFSTQDHK 170
Query: 173 LEENVEERERVTASGGEV--GRLNVFGGNEVGPLRCWPGGLCLSRSIGDTD--------- 221
N E+ER+ +GG V R+N G L +SR++GD D
Sbjct: 171 -PCNPVEKERIQNAGGSVMIQRVN--------------GSLAVSRALGDYDYKCVDGKGP 215
Query: 222 VGEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSD 258
+ + P P V ++ + +++A DGIWD +S++
Sbjct: 216 TEQLVSPEPEVYEIVRAEEDEFVVLACDGIWDVMSNE 252
>P2C_PARTE (P49444) Protein phosphatase 2C (EC 3.1.3.16) (PP2C)
Length = 300
Score = 66.6 bits (161), Expect = 1e-10
Identities = 67/255 (26%), Positives = 113/255 (44%), Gaps = 50/255 (19%)
Query: 54 DCHRVPGDSSTSFSVFAILDGHNGISAAIFAKENIINNVMSAIPQGVSREEWLQALPRAL 113
D H D SVF + DGH G A F +++ ++ ++ ++ Q AL
Sbjct: 38 DAHIHRHDIIQDVSVFGVFDGHGGREVAQFVEKHFVDELLK------NKNFKEQKFEEAL 91
Query: 114 VVAFVKTD-----MEFQKK----------GETSGTTATFVIIDGWTVTVASVGDSRCILD 158
F+K D E QK+ +G TA +I T+ VA+ GDSR +L
Sbjct: 92 KETFLKMDELLLTPEGQKELNQYKATDTDESYAGCTANVALIYKNTLYVANAGDSRSVL- 150
Query: 159 TQGGVVSLLTVDHRLEENVEERERVTASGGEV--GRLNVFGGNEVGPLRCWPGGLCLSRS 216
+ ++VDH+ +N EE+ R+ +GG V GR+N G L LSR+
Sbjct: 151 CRNNTNHDMSVDHK-PDNPEEKSRIERAGGFVSDGRVN--------------GNLNLSRA 195
Query: 217 IGDTDV---------GEYIVPIPHVKQVKLSNAGGRLIIASDGIWDTLSSDMAAKSCRGV 267
+GD + + I+ +P VK+ +L+ +++ DG+++TL+ K
Sbjct: 196 LGDLEYKRDNKLRSNEQLIIALPDVKKTELTPQDKFILMGCDGVFETLNHQELLKQVNST 255
Query: 268 --PAELAAKLVVKEA 280
A++ +L+ K A
Sbjct: 256 IGQAQVTEELLKKAA 270
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.135 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,297,497
Number of Sequences: 164201
Number of extensions: 2191902
Number of successful extensions: 5016
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 39
Number of HSP's that attempted gapping in prelim test: 4914
Number of HSP's gapped (non-prelim): 79
length of query: 423
length of database: 59,974,054
effective HSP length: 113
effective length of query: 310
effective length of database: 41,419,341
effective search space: 12839995710
effective search space used: 12839995710
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147963.9


