

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.5 - phase: 0
(459 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
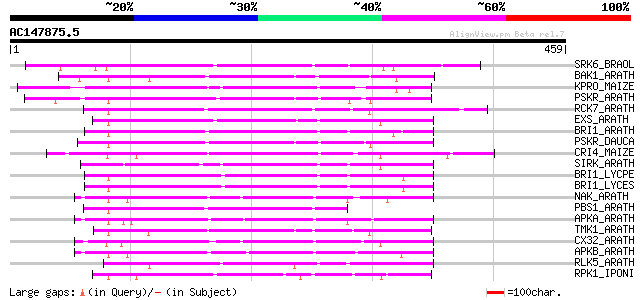
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 192 2e-48
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 181 3e-45
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 176 1e-43
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 175 2e-43
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 174 4e-43
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 167 4e-41
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 167 7e-41
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 164 3e-40
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 164 6e-40
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 162 2e-39
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 158 2e-38
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 157 4e-38
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 157 7e-38
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 155 2e-37
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 155 2e-37
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 154 5e-37
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 154 6e-37
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 152 2e-36
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 144 4e-34
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 143 8e-34
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 192 bits (488), Expect = 2e-48
Identities = 128/400 (32%), Positives = 213/400 (53%), Gaps = 31/400 (7%)
Query: 14 GVTMFFIIFISCYFKKGIRRPQMTIFR----KRRKHVDNNVEVFMQSYNLSIARRYSYAE 69
GV++ ++ + C +K+ +R + + +R +++ N V S ++ E
Sbjct: 453 GVSVLLLLIMFCLWKRKQKRAKASAISIANTQRNQNLPMNEMVLSSKREFSGEYKFEELE 512
Query: 70 --------VKRITNSFR--DKLGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFINE 118
V + T +F +KLG GG+G+VYK L DG+++AVK ++++ G +EF+NE
Sbjct: 513 LPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNE 572
Query: 119 VASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQ 178
V I+R H+N+V +LG C E +++ LIYE++ SLD +++ + + N F
Sbjct: 573 VTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKL---NWNERFD 629
Query: 179 IAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPG 238
I G+ARGL YLHQ RI+H D+K NILLD+N PKISDFG+A+I + +++ +
Sbjct: 630 ITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMK 689
Query: 239 TRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIY 298
GT GYM+PE +G S KSDV+S+G+++LE++ G+KN + + E +++
Sbjct: 690 VVGTYGYMSPEY--AMYGIFSEKSDVFSFGVIVLEIVSGKKN-RGFYNLDYENDLLSYVW 746
Query: 299 KDLEQGNDL-------LNSLTISEE--ENDMVKKITMVSLWCIQTNPLDRPPMNKVIEML 349
++G L ++SL+ + V K + L C+Q RP M+ V+ M
Sbjct: 747 SRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMF 806
Query: 350 QGPLSSVTFPPKPVLFSPKRPPLQLSNMSSSDWQETNSIT 389
+ + PKP + +R P +L SS E S T
Sbjct: 807 GSEATEIP-QPKPPGYCVRRSPYELDPSSSWQCDENESWT 845
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 181 bits (460), Expect = 3e-45
Identities = 115/322 (35%), Positives = 183/322 (56%), Gaps = 17/322 (5%)
Query: 41 KRRKHVDNNVEVFMQS---YNLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLT 95
+R+K D+ +V + +L +R+S E++ +++F +K LG GG+G VYK L
Sbjct: 250 RRKKPQDHFFDVPAEEDPEVHLGQLKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLA 309
Query: 96 DGRQVAVKVINESKGNGEE--FINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKG 153
DG VAVK + E + G E F EV IS H N++ L G+C +R L+Y +M G
Sbjct: 310 DGTLVAVKRLKEERTQGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANG 369
Query: 154 SLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDEN 213
S+ + + P++ D +IA+G ARGL YLH C +I+H D+K NILLDE
Sbjct: 370 SVASCLRER--PESQPPLDWPKRQRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEE 427
Query: 214 FCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILE 273
F + DFGLAK+ D+ V+ RGTIG++APE S G S K+DV+ YG+++LE
Sbjct: 428 FEAVVGDFGLAKLMDYKDTHVT-TAVRGTIGHIAPEYLST--GKSSEKTDVFGYGVMLLE 484
Query: 274 MIGGRKNYQTGGSCT-SEMYFPDWIYKDLEQGNDLLNSLTISEEEN---DMVKKITMVSL 329
+I G++ + ++ DW+ K L + L + + + N + V+++ V+L
Sbjct: 485 LITGQRAFDLARLANDDDVMLLDWV-KGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVAL 543
Query: 330 WCIQTNPLDRPPMNKVIEMLQG 351
C Q++P++RP M++V+ ML+G
Sbjct: 544 LCTQSSPMERPKMSEVVRMLEG 565
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 176 bits (446), Expect = 1e-43
Identities = 119/361 (32%), Positives = 188/361 (51%), Gaps = 43/361 (11%)
Query: 7 GASVAGFGVTMFFIIFISCY-FKKGIRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRY 65
G A F V + FI F + K+ +R ++ K K + +N RRY
Sbjct: 477 GFIAAFFVVEVSFISFAWFFVLKRELRPSELWASEKGYKAMTSNF------------RRY 524
Query: 66 SYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEEFINEVASISRT 125
SY E+ + T F+ +LG G G VYK L D R VAVK + + E F E++ I R
Sbjct: 525 SYRELVKATRKFKVELGRGESGTVYKGVLEDDRHVAVKKLENVRQGKEVFQAELSVIGRI 584
Query: 126 SHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIAR 185
+HMN+V + G+C E + R L+ E++ GSL ++ G + + D + F IA+G+A+
Sbjct: 585 NHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILFSEG-GNILLDWEGR--FNIALGVAK 641
Query: 186 GLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGY 245
GL YLH C ++H D+KP+NILLD+ F PKI+DFGL K+ S ++ RGT+GY
Sbjct: 642 GLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLGY 701
Query: 246 MAPEVFSRAFGGVSYKSDVYSYGMLILEMI-GGRKNYQTGGSCTSEMYFPDWIYKDLEQG 304
+APE S ++ K DVYSYG+++LE++ G R + GG+ D ++ L +
Sbjct: 702 IAPEWVSSL--PITAKVDVYSYGVVLLELLTGTRVSELVGGT--------DEVHSMLRKL 751
Query: 305 NDLLNSLTISEEEN--------------DMVKKITMVSL--WCIQTNPLDRPPMNKVIEM 348
+L++ EE++ + V+ T++ L C++ + RP M ++
Sbjct: 752 VRMLSAKLEGEEQSWIDGYLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAVQT 811
Query: 349 L 349
L
Sbjct: 812 L 812
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 175 bits (443), Expect = 2e-43
Identities = 115/348 (33%), Positives = 191/348 (54%), Gaps = 23/348 (6%)
Query: 13 FGVTMFFIIFISCYFKKGIRRPQM----TIFRKRRKHVDNNVEVFMQSYNLSIARRYSYA 68
F +T+ +I + + G P++ ++ RK + + + V QS + + SY
Sbjct: 670 FLLTLLSLIVLRARRRSGEVDPEIEESESMNRKELGEIGSKLVVLFQSND----KELSYD 725
Query: 69 EVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEVASISRT 125
++ TNSF +G GG+G+VYKA+L DG++VA+K ++ G E EF EV ++SR
Sbjct: 726 DLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRA 785
Query: 126 SHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIAR 185
H N+V L G+C+ N R LIY +M GSLD ++++ A+ T +IA G A+
Sbjct: 786 QHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKW--KTRLRIAQGAAK 843
Query: 186 GLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGY 245
GL YLH+GC ILH DIK NILLDENF ++DFGLA++ ++ VS GT+GY
Sbjct: 844 GLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVS-TDLVGTLGY 902
Query: 246 MAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK--NYQTGGSCTSEMYFPDWI--YKDL 301
+ PE + +YK DVYS+G+++LE++ ++ + C + W+ K
Sbjct: 903 IPPEYGQASV--ATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLI---SWVVKMKHE 957
Query: 302 EQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEML 349
+ +++ + L S+E + + ++ ++ C+ NP RP +++ L
Sbjct: 958 SRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 174 bits (441), Expect = 4e-43
Identities = 113/341 (33%), Positives = 179/341 (52%), Gaps = 15/341 (4%)
Query: 62 ARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQV-AVKVINESKGNG-EEFIN 117
A+ +++ E+ T +FR LG GG+G V+K ++ QV A+K ++ + G EF+
Sbjct: 88 AQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVV 147
Query: 118 EVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLF 177
EV ++S H N+V L+G+C E ++R L+YE+MP+GSL+ ++ P D NT
Sbjct: 148 EVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHV--LPSGKKPLDWNTRM 205
Query: 178 QIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIP 237
+IA G ARGLEYLH + +++ D+K NILL E++ PK+SDFGLAK+ D
Sbjct: 206 KIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVST 265
Query: 238 GTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDW- 296
GT GY AP+ G +++KSD+YS+G+++LE+I GRK + W
Sbjct: 266 RVMGTYGYCAPDY--AMTGQLTFKSDIYSFGVVLLELITGRKAID-NTKTRKDQNLVGWA 322
Query: 297 --IYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQGPLS 354
++KD +++ L + + + +S C+Q P RP ++ V+ L S
Sbjct: 323 RPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLAS 382
Query: 355 SVTFPPKPVLFSPKRPPLQLSNMSSSDWQETNSITTETELE 395
S P P S K P + D ++ + ETE E
Sbjct: 383 SKYDPNSPSSSSGKNPSF---HRDRDDEEKRPHLVKETECE 420
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 167 bits (424), Expect = 4e-41
Identities = 101/288 (35%), Positives = 165/288 (57%), Gaps = 12/288 (4%)
Query: 69 EVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEVASISRT 125
++ T+ F K +G GG+G VYKA L + VAVK ++E+K G EF+ E+ ++ +
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKV 968
Query: 126 SHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY-KSGFPDAVCDCDSNTLFQIAIGIA 184
H N+VSLLGYC + ++ L+YE+M GSLD ++ ++G + + D + +IA+G A
Sbjct: 969 KHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVL---DWSKRLKIAVGAA 1025
Query: 185 RGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIG 244
RGL +LH G I+H DIK NILLD +F PK++DFGLA++ +S VS GT G
Sbjct: 1026 RGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTV-IAGTFG 1084
Query: 245 YMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQG 304
Y+ PE A + K DVYS+G+++LE++ G++ + W + + QG
Sbjct: 1085 YIPPEYGQSA--RATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQG 1142
Query: 305 N--DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQ 350
D+++ L +S + ++ +++ C+ P RP M V++ L+
Sbjct: 1143 KAVDVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 167 bits (422), Expect = 7e-41
Identities = 102/295 (34%), Positives = 170/295 (57%), Gaps = 14/295 (4%)
Query: 63 RRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEV 119
R+ ++A++ + TN F + +G GG+G VYKA L DG VA+K + G G+ EF+ E+
Sbjct: 869 RKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEM 928
Query: 120 ASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQI 179
+I + H N+V LLGYC ++R L+YEFM GSL+ ++ A + +T +I
Sbjct: 929 ETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDP--KKAGVKLNWSTRRKI 986
Query: 180 AIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGT 239
AIG ARGL +LH CS I+H D+K N+LLDEN ++SDFG+A++ D+ +S+
Sbjct: 987 AIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTL 1046
Query: 240 RGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYK 299
GT GY+ PE + S K DVYSYG+++LE++ G++ T + W+ +
Sbjct: 1047 AGTPGYVPPEYYQSF--RCSTKGDVYSYGVVLLELLTGKR--PTDSPDFGDNNLVGWVKQ 1102
Query: 300 DLE-QGNDLLNSLTISEE---ENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQ 350
+ + +D+ + + E+ E ++++ + V++ C+ RP M +V+ M +
Sbjct: 1103 HAKLRISDVFDPELMKEDPALEIELLQHL-KVAVACLDDRAWRRPTMVQVMAMFK 1156
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 164 bits (416), Expect = 3e-40
Identities = 101/299 (33%), Positives = 162/299 (53%), Gaps = 11/299 (3%)
Query: 57 YNLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGE- 113
+N S ++ + T+SF +G GG+G+VYKA+L DG +VA+K ++ G +
Sbjct: 723 HNKDSNNELSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDR 782
Query: 114 EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDS 173
EF EV ++SR H N+V LLGYC N + LIY +M GSLD ++++ D D
Sbjct: 783 EFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEK--VDGPPSLDW 840
Query: 174 NTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSI 233
T +IA G A GL YLHQ C ILH DIK NILL + F ++DFGLA++ D+
Sbjct: 841 KTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTH 900
Query: 234 VSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYF 293
V+ GT+GY+ PE + +YK DVYS+G+++LE++ GR+ S
Sbjct: 901 VT-TDLVGTLGYIPPEYGQASV--ATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLI 957
Query: 294 PDWI--YKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQ 350
W+ K ++ +++ + ++ + + + ++ C+ NP RP +++ L+
Sbjct: 958 -SWVLQMKTEKRESEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 164 bits (414), Expect = 6e-40
Identities = 113/381 (29%), Positives = 200/381 (51%), Gaps = 20/381 (5%)
Query: 31 IRRPQMTIFRKRRKHVDNNVEVFMQSYNLSIARRYSYAEVKRITNSFRD--KLGHGGYGV 88
+R + T + R+ ++ ++ M+ + A+ +SY E+++ T F + ++G G +
Sbjct: 461 LRLAKSTAYSFRKDNM--KIQPDMEDLKIRRAQEFSYEELEQATGGFSEDSQVGKGSFSC 518
Query: 89 VYKASLTDGRQVAVK---VINESKGNGEEFINEVASISRTSHMNIVSLLGYCYEANKRAL 145
V+K L DG VAVK ++ K + +EF NE+ +SR +H ++++LLGYC + ++R L
Sbjct: 519 VFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLL 578
Query: 146 IYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKP 205
+YEFM GSL + ++ P+ + IA+ ARG+EYLH ++H DIK
Sbjct: 579 VYEFMAHGSLYQHLHGKD-PNLKKRLNWARRVTIAVQAARGIEYLHGYACPPVIHRDIKS 637
Query: 206 QNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVY 265
NIL+DE+ +++DFGL+ + + GT+GY+ PE + + ++ KSDVY
Sbjct: 638 SNILIDEDHNARVADFGLSILGPADSGTPLSELPAGTLGYLDPEYYRLHY--LTTKSDVY 695
Query: 266 SYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQGN--DLLNSLTISEEENDMVKK 323
S+G+++LE++ GRK E +W ++ G+ +L+ + + + +KK
Sbjct: 696 SFGVVLLEILSGRKAIDMQ---FEEGNIVEWAVPLIKAGDIFAILDPVLSPPSDLEALKK 752
Query: 324 ITMVSLWCIQTNPLDRPPMNKVIEMLQGPLSSVTFPP---KPVLFSPKRPPLQLSNMSSS 380
I V+ C++ DRP M+KV L+ L+ + P +P+L P L S M
Sbjct: 753 IASVACKCVRMRGKDRPSMDKVTTALEHALALLMGSPCIEQPIL--PTEVVLGSSRMHKV 810
Query: 381 DWQETNSITTETELEEEPIEG 401
+N +E EL + +G
Sbjct: 811 SQMSSNHSCSENELADGEDQG 831
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 162 bits (410), Expect = 2e-39
Identities = 105/296 (35%), Positives = 163/296 (54%), Gaps = 13/296 (4%)
Query: 59 LSIARRY-SYAEVKRITNSFRDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNG-EEFI 116
L A+RY Y+EV ITN+F +G GG+G VY + +G QVAVKV++E G +EF
Sbjct: 557 LKTAKRYFKYSEVVNITNNFERVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFR 615
Query: 117 NEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTL 176
EV + R H N+ SL+GYC E N LIYE+M +L Y +G + +
Sbjct: 616 AEVDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGD--YLAGKRSFILSWEER-- 671
Query: 177 FQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSI 236
+I++ A+GLEYLH GC I+H D+KP NILL+E K++DFGL++ + S
Sbjct: 672 LKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQIS 731
Query: 237 PGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDW 296
G+IGY+ PE +S ++ KSDVYS G+++LE+I G+ S T +++ D
Sbjct: 732 TVVAGSIGYLDPEYYSTR--QMNEKSDVYSLGVVLLEVITGQP--AIASSKTEKVHISDH 787
Query: 297 IYKDLEQGN--DLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQ 350
+ L G+ +++ + K++ ++L C + RP M++V+ L+
Sbjct: 788 VRSILANGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELK 843
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 158 bits (400), Expect = 2e-38
Identities = 97/294 (32%), Positives = 160/294 (53%), Gaps = 12/294 (4%)
Query: 63 RRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEV 119
R+ ++A++ TN F + +G GG+G VYKA L DG VA+K + G G+ EF E+
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEM 933
Query: 120 ASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQI 179
+I + H N+V LLGYC +R L+YE+M GSL+ ++ + + +I
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARR--KI 991
Query: 180 AIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGT 239
AIG ARGL +LH C I+H D+K N+LLDEN ++SDFG+A++ D+ +S+
Sbjct: 992 AIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTL 1051
Query: 240 RGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYK 299
GT GY+ PE + S K DVYSYG+++LE++ G++ T + + W+
Sbjct: 1052 AGTPGYVPPEYYQSF--RCSTKGDVYSYGVVLLELLTGKQ--PTDSADFGDNNLVGWVKL 1107
Query: 300 DLE-QGNDLLNSLTISEEENDMVKKI--TMVSLWCIQTNPLDRPPMNKVIEMLQ 350
+ + D+ + + E+ + ++ + V+ C+ RP M +V+ M +
Sbjct: 1108 HAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFK 1161
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 157 bits (398), Expect = 4e-38
Identities = 97/294 (32%), Positives = 160/294 (53%), Gaps = 12/294 (4%)
Query: 63 RRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGE-EFINEV 119
R+ ++A++ TN F + +G GG+G VYKA L DG VA+K + G G+ EF E+
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEM 933
Query: 120 ASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQI 179
+I + H N+V LLGYC +R L+YE+M GSL+ ++ + + +I
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARR--KI 991
Query: 180 AIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGT 239
AIG ARGL +LH C I+H D+K N+LLDEN ++SDFG+A++ D+ +S+
Sbjct: 992 AIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTL 1051
Query: 240 RGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYK 299
GT GY+ PE + S K DVYSYG+++LE++ G++ T + + W+
Sbjct: 1052 AGTPGYVPPEYYQSF--RCSTKGDVYSYGVVLLELLTGKQ--PTDSADFGDNNLVGWVKL 1107
Query: 300 DLE-QGNDLLNSLTISEEENDMVKKI--TMVSLWCIQTNPLDRPPMNKVIEMLQ 350
+ + D+ + + E+ + ++ + V+ C+ RP M +V+ M +
Sbjct: 1108 HAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFK 1161
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 157 bits (396), Expect = 7e-38
Identities = 107/317 (33%), Positives = 166/317 (51%), Gaps = 33/317 (10%)
Query: 54 MQSYNLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTD----------GRQVA 101
+Q+ NL + +S +E+K T +FR +G GG+G V+K + + G +A
Sbjct: 48 LQNANL---KNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIA 104
Query: 102 VKVINESKGNGE-EFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
VK +N+ G E++ E+ + + H N+V L+GYC E R L+YEFM +GSL+ ++
Sbjct: 105 VKRLNQEGFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLF 164
Query: 161 KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISD 220
+ G NT ++A+G ARGL +LH ++++ D K NILLD N+ K+SD
Sbjct: 165 RRG--TFYQPLSWNTRVRMALGAARGLAFLH-NAQPQVIYRDFKASNILLDSNYNAKLSD 221
Query: 221 FGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR-- 278
FGLA+ M D+ GT GY APE A G +S KSDVYS+G+++LE++ GR
Sbjct: 222 FGLARDGPMGDNSHVSTRVMGTQGYAAPEYL--ATGHLSVKSDVYSFGVVLLELLSGRRA 279
Query: 279 --KNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSL---TISEEENDMVKKITMVSLWCIQ 333
KN G E DW L LL + + KI +++L CI
Sbjct: 280 IDKNQPVG-----EHNLVDWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCIS 334
Query: 334 TNPLDRPPMNKVIEMLQ 350
+ RP MN++++ ++
Sbjct: 335 IDAKSRPTMNEIVKTME 351
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 155 bits (392), Expect = 2e-37
Identities = 93/222 (41%), Positives = 131/222 (58%), Gaps = 8/222 (3%)
Query: 62 ARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASL-TDGRQVAVKVINESKGNGE-EFIN 117
A +++ E+ T +F LG GG+G VYK L + G+ VAVK ++ + G EF+
Sbjct: 71 AHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLV 130
Query: 118 EVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLF 177
EV +S H N+V+L+GYC + ++R L+YEFMP GSL+ ++ P D N
Sbjct: 131 EVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHD--LPPDKEALDWNMRM 188
Query: 178 QIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIP 237
+IA G A+GLE+LH + +++ D K NILLDE F PK+SDFGLAK+ D
Sbjct: 189 KIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVST 248
Query: 238 GTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK 279
GT GY APE G ++ KSDVYS+G++ LE+I GRK
Sbjct: 249 RVMGTYGYCAPEY--AMTGQLTVKSDVYSFGVVFLELITGRK 288
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 155 bits (392), Expect = 2e-37
Identities = 109/314 (34%), Positives = 170/314 (53%), Gaps = 27/314 (8%)
Query: 54 MQSYNLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKA-----SLTDGRQ-----VA 101
+QS NL + +S+AE+K T +FR LG GG+G V+K SLT R +A
Sbjct: 48 LQSPNL---KSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIA 104
Query: 102 VKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
VK +N+ G +E++ EV + + SH ++V L+GYC E R L+YEFMP+GSL+ ++
Sbjct: 105 VKKLNQDGWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF 164
Query: 161 KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISD 220
+ G ++A+G A+GL +LH +R+++ D K NILLD + K+SD
Sbjct: 165 RRGL--YFQPLSWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSD 221
Query: 221 FGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGR-- 278
FGLAK + D GT GY APE A G ++ KSDVYS+G+++LE++ GR
Sbjct: 222 FGLAKDGPIGDKSHVSTRVMGTHGYAAPEYL--ATGHLTTKSDVYSFGVVLLELLSGRRA 279
Query: 279 --KNYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNP 336
KN +G E P + K + ++++ + + K+ +SL C+ T
Sbjct: 280 VDKNRPSGERNLVEWAKPYLVNK--RKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEI 337
Query: 337 LDRPPMNKVIEMLQ 350
RP M++V+ L+
Sbjct: 338 KLRPNMSEVVSHLE 351
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 154 bits (389), Expect = 5e-37
Identities = 91/290 (31%), Positives = 155/290 (53%), Gaps = 15/290 (5%)
Query: 70 VKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVKVINESKGNGE---EFINEVASISR 124
++ +TN+F LG GG+GVVYK L DG ++AVK + G+ EF +E+A +++
Sbjct: 581 LRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTK 640
Query: 125 TSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIA 184
H ++V+LLGYC + N++ L+YE+MP+G+L + +++ + + +A+ +A
Sbjct: 641 VRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWS-EEGLKPLLWKQRLTLALDVA 699
Query: 185 RGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGTRGTIG 244
RG+EYLH +H D+KP NILL ++ K++DFGL ++ + GT G
Sbjct: 700 RGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIE-TRIAGTFG 758
Query: 245 YMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDW-----IYK 299
Y+APE G V+ K DVYS+G++++E+I GRK+ ++ W I K
Sbjct: 759 YLAPEY--AVTGRVTTKVDVYSFGVILMELITGRKSLDE-SQPEESIHLVSWFKRMYINK 815
Query: 300 DLEQGNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEML 349
+ + ++ + EE V + ++ C P RP M + +L
Sbjct: 816 EASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNIL 865
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 154 bits (388), Expect = 6e-37
Identities = 101/313 (32%), Positives = 166/313 (52%), Gaps = 28/313 (8%)
Query: 54 MQSYNLSIARRYSYAEVKRITNSFR--DKLGHGGYGVVYK----------ASLTDGRQVA 101
++S NL + Y++ ++K T +F+ LG GG+G VY+ + + G VA
Sbjct: 66 LESPNLKV---YNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVA 122
Query: 102 VKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
+K +N G E+ +EV + SH N+V LLGYC E + L+YEFMPKGSL+ ++
Sbjct: 123 IKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLF 182
Query: 161 KSGFPDAVCDCDSNTLFQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISD 220
+ P + +I IG ARGL +LH +++ D K NILLD N+ K+SD
Sbjct: 183 RRNDP-----FPWDLRIKIVIGAARGLAFLH-SLQREVIYRDFKASNILLDSNYDAKLSD 236
Query: 221 FGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKN 280
FGLAK+ ++ GT GY APE A G + KSDV+++G+++LE++ G
Sbjct: 237 FGLAKLGPADEKSHVTTRIMGTYGYAAPEYM--ATGHLYVKSDVFAFGVVLLEIMTGLTA 294
Query: 281 YQTGGSCTSEMYFPDWIYKDLEQGN---DLLNSLTISEEENDMVKKITMVSLWCIQTNPL 337
+ T E DW+ +L + +++ + + ++ ++L CI+ +P
Sbjct: 295 HNTKRPRGQES-LVDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPK 353
Query: 338 DRPPMNKVIEMLQ 350
+RP M +V+E+L+
Sbjct: 354 NRPHMKEVVEVLE 366
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 152 bits (383), Expect = 2e-36
Identities = 107/314 (34%), Positives = 167/314 (53%), Gaps = 27/314 (8%)
Query: 54 MQSYNLSIARRYSYAEVKRITNSFRDK--LGHGGYGVVYKASLTD----------GRQVA 101
+QS NL + +++AE+K T +FR LG GG+G V+K + + G +A
Sbjct: 49 LQSPNL---KSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIA 105
Query: 102 VKVINESKGNG-EEFINEVASISRTSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIY 160
VK +N+ G +E++ EV + + SH N+V L+GYC E R L+YEFMP+GSL+ ++
Sbjct: 106 VKKLNQDGWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF 165
Query: 161 KSGFPDAVCDCDSNTL-FQIAIGIARGLEYLHQGCSSRILHLDIKPQNILLDENFCPKIS 219
+ G + S TL ++A+G A+GL +LH +S +++ D K NILLD + K+S
Sbjct: 166 RRG---SYFQPLSWTLRLKVALGAAKGLAFLHNAETS-VIYRDFKTSNILLDSEYNAKLS 221
Query: 220 DFGLAKICQMNDSIVSIPGTRGTIGYMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRK 279
DFGLAK D GT GY APE A G ++ KSDVYSYG+++LE++ GR+
Sbjct: 222 DFGLAKDGPTGDKSHVSTRIMGTYGYAAPEYL--ATGHLTTKSDVYSYGVVLLEVLSGRR 279
Query: 280 NYQTGGSCTSEMYFPDWIYKDLEQGNDLLNSLTISEEENDMVK---KITMVSLWCIQTNP 336
E +W L L + ++ ++ K+ ++L C+
Sbjct: 280 AVDKNRP-PGEQKLVEWARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEI 338
Query: 337 LDRPPMNKVIEMLQ 350
RP MN+V+ L+
Sbjct: 339 KLRPNMNEVVSHLE 352
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 144 bits (364), Expect = 4e-34
Identities = 94/287 (32%), Positives = 148/287 (50%), Gaps = 21/287 (7%)
Query: 78 RDKLGHGGYGVVYKASLTDGRQVAVKVINESKGNGEE-----------FINEVASISRTS 126
++ +G G G VYK L G VAVK +N+S G++ F EV ++
Sbjct: 686 KNVIGFGSSGKVYKVELRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTIR 745
Query: 127 HMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIARG 186
H +IV L C + + L+YE+MP GSL ++ V +IA+ A G
Sbjct: 746 HKSIVRLWCCCSSGDCKLLVYEYMPNGSLADVLHGDRKGGVVLGWPER--LRIALDAAEG 803
Query: 187 LEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIV--SIPGTRGTIG 244
L YLH C I+H D+K NILLD ++ K++DFG+AK+ QM+ S ++ G G+ G
Sbjct: 804 LSYLHHDCVPPIVHRDVKSSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMSGIAGSCG 863
Query: 245 YMAPEVFSRAFGGVSYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDLEQ- 303
Y+APE V+ KSD+YS+G+++LE++ G+ Q S + W+ L++
Sbjct: 864 YIAPEYVYTL--RVNEKSDIYSFGVVLLELVTGK---QPTDSELGDKDMAKWVCTALDKC 918
Query: 304 GNDLLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEMLQ 350
G + + + + + + K+ + L C PL+RP M KV+ MLQ
Sbjct: 919 GLEPVIDPKLDLKFKEEISKVIHIGLLCTSPLPLNRPSMRKVVIMLQ 965
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 143 bits (361), Expect = 8e-34
Identities = 102/296 (34%), Positives = 150/296 (50%), Gaps = 26/296 (8%)
Query: 69 EVKRITNSFRDK--LGHGGYGVVYKASLTDGRQVAVK--VINESKGNGEEFINEVASISR 124
+V T + DK +G G +G +YKA+L+ + AVK V K + E+ +I +
Sbjct: 808 KVLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGK 867
Query: 125 TSHMNIVSLLGYCYEANKRALIYEFMPKGSLDKFIYKSGFPDAVCDCDSNTLFQIAIGIA 184
H N++ L + ++Y +M GSL ++++ P + D +T IA+G A
Sbjct: 868 VRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPL---DWSTRHNIAVGTA 924
Query: 185 RGLEYLHQGCSSRILHLDIKPQNILLDENFCPKISDFGLAKICQMNDSIVSIPGT--RGT 242
GL YLH C I+H DIKP NILLD + P ISDFG+AK+ ++ S SIP +GT
Sbjct: 925 HGLAYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKL--LDQSATSIPSNTVQGT 982
Query: 243 IGYMAPEVFSRAFGGV-SYKSDVYSYGMLILEMIGGRKNYQTGGSCTSEMYFPDWIYKDL 301
IGYMAPE AF V S +SDVYSYG+++LE+I +K S E W+
Sbjct: 983 IGYMAPE---NAFTTVKSRESDVYSYGVVLLELITRKKALDP--SFNGETDIVGWVRSVW 1037
Query: 302 EQGND--------LLNSLTISEEENDMVKKITMVSLWCIQTNPLDRPPMNKVIEML 349
Q + LL+ L I + V + ++L C + RP M V++ L
Sbjct: 1038 TQTGEIQKIVDPSLLDEL-IDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 50,749,334
Number of Sequences: 164201
Number of extensions: 2140675
Number of successful extensions: 8435
Number of sequences better than 10.0: 1646
Number of HSP's better than 10.0 without gapping: 978
Number of HSP's successfully gapped in prelim test: 668
Number of HSP's that attempted gapping in prelim test: 5428
Number of HSP's gapped (non-prelim): 1849
length of query: 459
length of database: 59,974,054
effective HSP length: 114
effective length of query: 345
effective length of database: 41,255,140
effective search space: 14233023300
effective search space used: 14233023300
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147875.5


