

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.4 + phase: 0
(687 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
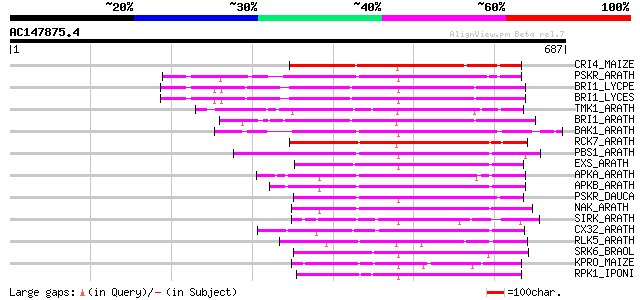
Score E
Sequences producing significant alignments: (bits) Value
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 216 2e-55
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 204 6e-52
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 204 8e-52
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 202 2e-51
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 199 2e-50
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 199 2e-50
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 199 3e-50
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 197 8e-50
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 197 8e-50
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 193 1e-48
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 192 3e-48
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 189 2e-47
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 187 8e-47
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 180 1e-44
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 179 2e-44
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 179 3e-44
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 174 5e-43
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 174 9e-43
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 164 7e-40
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 157 9e-38
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 216 bits (550), Expect = 2e-55
Identities = 125/294 (42%), Positives = 188/294 (63%), Gaps = 13/294 (4%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNF--KRVAQFMN 404
Q F+YEELE+AT F ++G+G F V+KG L+DG VVAVKR +++ K +F N
Sbjct: 491 QEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHN 550
Query: 405 EVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHG-DRSSSCLLPWSVRL 463
E+++L+RL H +L+ L G S E LLVYE++++G++ HLHG D + L W+ R+
Sbjct: 551 ELDLLSRLNHAHLLNLLGYCEDGS-ERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRV 609
Query: 464 DIALETAEALAYLHA---SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDV-THVST 519
IA++ A + YLH V+HRD+KS+NIL+DE + +VADFGLS L P D T +S
Sbjct: 610 TIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSE 669
Query: 520 APQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNK 579
P GT GY+DPEYY+ + LT KSDVYSFGVVL+E++S +A+D+ + N+ AV
Sbjct: 670 LPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDM--QFEEGNIVEWAVPL 727
Query: 580 IQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
I++ +++ ++DP L D + +A +A +C++ + RPSMD++ L
Sbjct: 728 IKAGDIFAILDPVLSPPSDLEALK---KIASVACKCVRMRGKDRPSMDKVTTAL 778
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 204 bits (519), Expect = 6e-52
Identities = 158/461 (34%), Positives = 229/461 (49%), Gaps = 48/461 (10%)
Query: 190 LEEQANRFAGNR--SLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGF--DSDENEHI 245
L+ NR +G+ SL + + F+V YNN I SGGQ F S E+ H+
Sbjct: 576 LDLSNNRLSGSIPVSLQQLSFLSKFSVAYNN-----LSGVIPSGGQFQTFPNSSFESNHL 630
Query: 246 CICGNGLCPSGNSS--------TNGGLIGGVVG-GVAALCLLGFVACFVVRRRRKNAKKP 296
C C G S + GG IG +G ++ LL ++ V+R RR++ +
Sbjct: 631 CGEHRFPCSEGTESALIKRSRRSRGGDIGMAIGIAFGSVFLLTLLSLIVLRARRRSGEV- 689
Query: 297 ISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEE 356
D + S + + G + S + F + +Y++L +
Sbjct: 690 ---DPEIEESESMNRKELGEIGSK-----------------LVVLFQSNDKELSYDDLLD 729
Query: 357 ATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKN 416
+TN+F + +G GGFG VYK L DG+ VA+K+ + +F EVE L+R +H N
Sbjct: 730 STNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRAQHPN 789
Query: 417 LVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAY 475
LV L G C K+ R LL+Y Y+ NG++ LH LL W RL IA A+ L Y
Sbjct: 790 LVLLRGFCFYKNDR--LLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLY 847
Query: 476 LHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEY 532
LH ++HRD+KS+NILLDE F+ +ADFGL+RL THVST GT GY+ PEY
Sbjct: 848 LHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTDLVGTLGYIPPEY 907
Query: 533 YQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPN 592
Q T K DVYSFGVVL+EL++ + VD+ + + +L + V ++ DP
Sbjct: 908 GQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVFDP- 966
Query: 593 LGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
L Y K+N + V E+A CL + RP+ ++V L
Sbjct: 967 LIYSKENDKEMF--RVLEIACLCLSENPKQRPTTQQLVSWL 1005
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 204 bits (518), Expect = 8e-52
Identities = 151/473 (31%), Positives = 235/473 (48%), Gaps = 41/473 (8%)
Query: 187 VPILEEQANRFAGN--RSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEH 244
V IL+ NRF G SL L+ +++ NN I F +
Sbjct: 713 VAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNN-----LSGMIPESAPFDTFPDYRFAN 767
Query: 245 ICICGNGL---CPSGNSST----------NGGLIGGVVGGV--AALCLLGFVACFVVRRR 289
+CG L C SG S L G V G+ + C+ G + + ++
Sbjct: 768 NSLCGYPLPLPCSSGPKSDANQHQKSHRRQASLAGSVAMGLLFSLFCIFGLIIVAIETKK 827
Query: 290 RKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVF 349
R+ KK + + YM S ++ N+ ++ + SI K ++
Sbjct: 828 RRR-KKEAALEAYMDGHSHSATANSAWKFTSAREALSINLAAFEKP----------LRKL 876
Query: 350 TYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEIL 409
T+ +L EATN FH +G GGFG VYK LKDG VVA+K+ + + +F E+E +
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETI 936
Query: 410 ARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALET 469
+++H+NLV L G K E LLVYEY+ G++ D LH + + L W R IA+
Sbjct: 937 GKIKHRNLVPLLG-YCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAIGA 995
Query: 470 AEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAP-QGTP 525
A LA+LH + ++HRD+KS+N+LLDE +V+DFG++RL TH+S + GTP
Sbjct: 996 ARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 1055
Query: 526 GYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQEL 585
GYV PEYYQ ++ + K DVYS+GVVL+EL++ Q D + D NL V ++
Sbjct: 1056 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGW-VKLHAKGKI 1113
Query: 586 YDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKS 638
D+ D L ++D S++ ++A CL + RP+M +++ + + I++
Sbjct: 1114 TDVFDREL-LKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQA 1165
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 202 bits (515), Expect = 2e-51
Identities = 151/473 (31%), Positives = 234/473 (48%), Gaps = 41/473 (8%)
Query: 187 VPILEEQANRFAGN--RSLLREVLMEGFNVNYNNPFENDCLECISSGGQQCGFDSDENEH 244
V IL+ NRF G SL L+ +++ NN I F +
Sbjct: 713 VAILDLSYNRFNGTIPNSLTSLTLLGEIDLSNNN-----LSGMIPESAPFDTFPDYRFAN 767
Query: 245 ICICGNGL---CPSGNSST----------NGGLIGGVVGGV--AALCLLGFVACFVVRRR 289
+CG L C SG S L G V G+ + C+ G + + ++
Sbjct: 768 NSLCGYPLPIPCSSGPKSDANQHQKSHRRQASLAGSVAMGLLFSLFCIFGLIIVAIETKK 827
Query: 290 RKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVF 349
R+ KK + + YM S ++ N+ ++ + SI K ++
Sbjct: 828 RRR-KKEAALEAYMDGHSHSATANSAWKFTSAREALSINLAAFEKP----------LRKL 876
Query: 350 TYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEIL 409
T+ +L EATN FH +G GGFG VYK LKDG VVA+K+ + + +F E+E +
Sbjct: 877 TFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEMETI 936
Query: 410 ARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALET 469
+++H+NLV L G K E LLVYEY+ G++ D LH + L W R IA+
Sbjct: 937 GKIKHRNLVPLLG-YCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAIGA 995
Query: 470 AEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAP-QGTP 525
A LA+LH + ++HRD+KS+N+LLDE +V+DFG++RL TH+S + GTP
Sbjct: 996 ARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTP 1055
Query: 526 GYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQEL 585
GYV PEYYQ ++ + K DVYS+GVVL+EL++ Q D + D NL V ++
Sbjct: 1056 GYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD-SADFGDNNLVGW-VKLHAKGKI 1113
Query: 586 YDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKS 638
D+ D L ++D S++ ++A CL + RP+M +++ + + I++
Sbjct: 1114 TDVFDREL-LKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQA 1165
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 199 bits (507), Expect = 2e-50
Identities = 145/429 (33%), Positives = 222/429 (50%), Gaps = 41/429 (9%)
Query: 230 SGGQQCGFDSDENEHICICGNGLCPSGNSSTNGGLIGGVVGGVAALCLLGF-VACFVVRR 288
SGG G + D++ +S+ G ++G V+GG+ ++ L+G V C+ +R
Sbjct: 459 SGGSGSGINGDKDRR---------GMKSSTFIGIIVGSVLGGLLSIFLIGLLVFCWYKKR 509
Query: 289 RRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQV 348
+++ + SN + + P + S + +T SS S+ S T T+P + G +
Sbjct: 510 QKRFSGSESSNAVVVHPRHSGSDNESVKIT-VAGSSVSVGGI--SDTYTLPGTSEVGDNI 566
Query: 349 ---------FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNF--K 397
+ + L TNNF + LG GGFG VYKG+L DG +AVKR K
Sbjct: 567 QMVEAGNMLISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGK 626
Query: 398 RVAQFMNEVEILARLRHKNLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCL 456
A+F +E+ +L ++RH++LVTL G C E LLVYEY+ GT++ HL + S L
Sbjct: 627 GFAEFKSEIAVLTKVRHRHLVTLLGYCLD--GNEKLLVYEYMPQGTLSRHLF-EWSEEGL 683
Query: 457 LP--WSVRLDIALETAEALAYLHA---SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFP 511
P W RL +AL+ A + YLH +HRD+K +NILL + KVADFGL RL P
Sbjct: 684 KPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAP 743
Query: 512 NDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVN 571
+ T GT GY+ PEY ++T K DVYSFGV+L+ELI+ +++D ++ ++
Sbjct: 744 EGKGSIETRIAGTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIH 803
Query: 572 LAN----MAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMD 627
L + M +NK S +D + +++ T VAELA C ++ RP M
Sbjct: 804 LVSWFKRMYINKEAS--FKKAIDTTIDLDEETLASVHT--VAELAGHCCAREPYQRPDMG 859
Query: 628 EIVEVLRAI 636
V +L ++
Sbjct: 860 HAVNILSSL 868
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 199 bits (507), Expect = 2e-50
Identities = 130/401 (32%), Positives = 216/401 (53%), Gaps = 20/401 (4%)
Query: 260 TNGGLIGGVVGGVAALCLLGFVACFVV-----RRRRKNAKKPISNDLYMPPSSTTSGTNT 314
++G + G VA L FV F + R++ KK ++Y N+
Sbjct: 784 SHGRRPASLAGSVAMGLLFSFVCIFGLILVGREMRKRRRKKEAELEMYAEGHG-----NS 838
Query: 315 GTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGT 374
G T+ N++ + + + + +F ++ T+ +L +ATN FH +G GGFG
Sbjct: 839 GDRTAN-NTNWKLTGVKEALSINLA-AFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGD 896
Query: 375 VYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLV 434
VYK LKDG VA+K+ + + +FM E+E + +++H+NLV L G K E LLV
Sbjct: 897 VYKAILKDGSAVAIKKLIHVSGQGDREFMAEMETIGKIKHRNLVPLLG-YCKVGDERLLV 955
Query: 435 YEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLH---ASDVMHRDVKSNNI 491
YE++ G++ D LH + + L WS R IA+ +A LA+LH + ++HRD+KS+N+
Sbjct: 956 YEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHIIHRDMKSSNV 1015
Query: 492 LLDEKFHVKVADFGLSRLFPNDVTHVSTAP-QGTPGYVDPEYYQCYQLTDKSDVYSFGVV 550
LLDE +V+DFG++RL TH+S + GTPGYV PEYYQ ++ + K DVYS+GVV
Sbjct: 1016 LLDENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVV 1075
Query: 551 LVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAE 610
L+EL++ + D + D NL V + + D+ DP L ++D +++ +
Sbjct: 1076 LLELLTGKRPTD-SPDFGDNNLVGW-VKQHAKLRISDVFDPEL-MKEDPALEIELLQHLK 1132
Query: 611 LAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDV 651
+A CL + RP+M +++ + + I++ +S + +
Sbjct: 1133 VAVACLDDRAWRRPTMVQVMAMFKEIQAGSGIDSQSTIRSI 1173
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 199 bits (505), Expect = 3e-50
Identities = 153/439 (34%), Positives = 219/439 (49%), Gaps = 59/439 (13%)
Query: 254 PSGNSSTNGGLIGGVVGGVAALCLLGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTN 313
P+G++ G + GGV G A L + +A RR KKP + + P+ +
Sbjct: 217 PAGSNRITGAIAGGVAAGAALLFAVPAIALAWWRR-----KKP-QDHFFDVPAEEDPEVH 270
Query: 314 TGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFG 373
G L F+ EL+ A++NF LG GGFG
Sbjct: 271 LGQLKR-----------------------------FSLRELQVASDNFSNKNILGRGGFG 301
Query: 374 TVYKGDLKDGRVVAVKRHYESNFKR-VAQFMNEVEILARLRHKNLVTLYG-CTSKHSREL 431
VYKG L DG +VAVKR E + QF EVE+++ H+NL+ L G C + R
Sbjct: 302 KVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTER-- 359
Query: 432 LLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHAS---DVMHRDVKS 488
LLVY Y++NG+VA L S L W R IAL +A LAYLH ++HRDVK+
Sbjct: 360 LLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALGSARGLAYLHDHCDPKIIHRDVKA 419
Query: 489 NNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFG 548
NILLDE+F V DFGL++L THV+TA +GT G++ PEY + ++K+DV+ +G
Sbjct: 420 ANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTIGHIAPEYLSTGKSSEKTDVFGYG 479
Query: 549 VVLVELISSLQAVDITRHRN--DVNLANMAVNKIQSQELYDLVDPNL-GYEKDNSVKRMT 605
V+L+ELI+ +A D+ R N DV L + ++ ++L LVD +L G KD V+++
Sbjct: 480 VMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEKKLEALVDVDLQGNYKDEEVEQL- 538
Query: 606 TAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPETQESKVLDVVVRTDELVLLKKGP 665
++A C Q RP M E+V +L E + K E + +
Sbjct: 539 ---IQVALLCTQSSPMERPKMSEVVRMLEGDGLAERWEEWQK---------EEMFRQDFN 586
Query: 666 YPTSPDSVAEKWVSGSSTS 684
YPT +V+ W+ G STS
Sbjct: 587 YPTHHPAVS-GWIIGDSTS 604
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 197 bits (501), Expect = 8e-50
Identities = 123/300 (41%), Positives = 181/300 (60%), Gaps = 10/300 (3%)
Query: 347 QVFTYEELEEATNNFHTSKELGEGGFGTVYKGDL-KDGRVVAVKRHYESNFKRVAQFMNE 405
Q FT++EL EAT NF + LGEGGFG V+KG + K +VVA+K+ + + + +F+ E
Sbjct: 89 QTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVE 148
Query: 406 VEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDI 465
V L+ H NLV L G ++ + LL VYEY+ G++ DHLH S L W+ R+ I
Sbjct: 149 VLTLSLADHPNLVKLIGFCAEGDQRLL-VYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKI 207
Query: 466 ALETAEALAYLH---ASDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAP 521
A A L YLH V++RD+K +NILL E + K++DFGL+++ P+ D THVST
Sbjct: 208 AAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRV 267
Query: 522 QGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQ 581
GT GY P+Y QLT KSD+YSFGVVL+ELI+ +A+D T+ R D NL A +
Sbjct: 268 MGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWARPLFK 327
Query: 582 SQELY-DLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDE 640
+ + +VDP L + V+ + A+A ++ C+Q+Q +RP + ++V L + S +
Sbjct: 328 DRRNFPKMVDPLL--QGQYPVRGLYQALA-ISAMCVQEQPTMRPVVSDVVLALNFLASSK 384
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 197 bits (501), Expect = 8e-50
Identities = 143/397 (36%), Positives = 195/397 (49%), Gaps = 22/397 (5%)
Query: 278 LGFVACFVVRRRRKNAKKPISNDLYMPPSSTTSGTNTGTLTSTTNSSQSIPSYPSSKTST 337
+G +CF K SN S T N L S S + S +
Sbjct: 1 MGCFSCFDSSDDEKLNPVDESNHGQKKQSQPTVSNNISGLPSGGEKLSSKTNGGSKRELL 60
Query: 338 MPKSFY--FGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKD-GRVVAVKRHYES 394
+P+ F + EL AT NFH LGEGGFG VYKG L G+VVAVK+ +
Sbjct: 61 LPRDGLGQIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRN 120
Query: 395 NFKRVAQFMNEVEILARLRHKNLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSS 453
+ +F+ EV +L+ L H NLV L G C R LLVYE++ G++ DHLH
Sbjct: 121 GLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQR--LLVYEFMPLGSLEDHLHDLPPD 178
Query: 454 SCLLPWSVRLDIALETAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLF 510
L W++R+ IA A+ L +LH V++RD KS+NILLDE FH K++DFGL++L
Sbjct: 179 KEALDWNMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLG 238
Query: 511 PN-DVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRND 569
P D +HVST GT GY PEY QLT KSDVYSFGVV +ELI+ +A+D +
Sbjct: 239 PTGDKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGE 298
Query: 570 VNLANMAVNKIQSQELY-DLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDE 628
NL A + + L DP L K R +A C+Q+Q RP + +
Sbjct: 299 QNLVAWARPLFNDRRKFIKLADPRL---KGRFPTRALYQALAVASMCIQEQAATRPLIAD 355
Query: 629 IVEVLRAI--------KSDEPETQESKVLDVVVRTDE 657
+V L + K D ++ + ++ R D+
Sbjct: 356 VVTALSYLANQAYDPSKDDSRRNRDERGARLITRNDD 392
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 193 bits (491), Expect = 1e-48
Identities = 115/288 (39%), Positives = 168/288 (57%), Gaps = 8/288 (2%)
Query: 353 ELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILARL 412
++ EAT++F +G+GGFGTVYK L + VAVK+ E+ + +FM E+E L ++
Sbjct: 909 DIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKV 968
Query: 413 RHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEA 472
+H NLV+L G S S E LLVYEY+ NG++ L +L WS RL IA+ A
Sbjct: 969 KHPNLVSLLGYCS-FSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARG 1027
Query: 473 LAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGYVD 529
LA+LH ++HRD+K++NILLD F KVADFGL+RL +HVST GT GY+
Sbjct: 1028 LAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIAGTFGYIP 1087
Query: 530 PEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDIT-RHRNDVNLANMAVNKIQSQELYDL 588
PEY Q + T K DVYSFGV+L+EL++ + + NL A+ KI + D+
Sbjct: 1088 PEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAVDV 1147
Query: 589 VDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAI 636
+DP L ++K + ++A CL + RP+M ++++ L+ I
Sbjct: 1148 IDPLL---VSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALKEI 1192
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 192 bits (487), Expect = 3e-48
Identities = 131/351 (37%), Positives = 196/351 (55%), Gaps = 30/351 (8%)
Query: 306 SSTTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSK 365
S+ + G+L S +S PS P ++ + ++ F++ EL+ AT NF
Sbjct: 17 STKYDAKDIGSLGSKASSVSVRPS-PRTEGEILQSP---NLKSFSFAELKSATRNFRPDS 72
Query: 366 ELGEGGFGTVYKGDLKD----------GRVVAVKRHYESNFKRVAQFMNEVEILARLRHK 415
LGEGGFG V+KG + + G V+AVK+ + ++ +++ EV L + H+
Sbjct: 73 VLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQEWLAEVNYLGQFSHR 132
Query: 416 NLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALA 474
+LV L G C R LLVYE++ G++ +HL L W +RL +AL A+ LA
Sbjct: 133 HLVKLIGYCLEDEHR--LLVYEFMPRGSLENHLFRRGLYFQPLSWKLRLKVALGAAKGLA 190
Query: 475 YLHASD--VMHRDVKSNNILLDEKFHVKVADFGLSRLFP-NDVTHVSTAPQGTPGYVDPE 531
+LH+S+ V++RD K++NILLD +++ K++DFGL++ P D +HVST GT GY PE
Sbjct: 191 FLHSSETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTRVMGTHGYAAPE 250
Query: 532 YYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMA----VNKIQSQELYD 587
Y LT KSDVYSFGVVL+EL+S +AVD R + NL A VNK ++++
Sbjct: 251 YLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYLVNK---RKIFR 307
Query: 588 LVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKS 638
++D L +D VA L+ RCL + LRP+M E+V L I+S
Sbjct: 308 VIDNRL---QDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLEHIQS 355
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 189 bits (480), Expect = 2e-47
Identities = 125/332 (37%), Positives = 190/332 (56%), Gaps = 23/332 (6%)
Query: 322 NSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLK 381
+SS SI + P ++ + ++ FT+ EL+ AT NF LGEGGFG+V+KG +
Sbjct: 33 SSSVSIRTNPRTEGEILQSP---NLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWID 89
Query: 382 D----------GRVVAVKRHYESNFKRVAQFMNEVEILARLRHKNLVTLYG-CTSKHSRE 430
+ G V+AVK+ + ++ +++ EV L + H NLV L G C R
Sbjct: 90 EQTLTASKPGTGVVIAVKKLNQDGWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHR- 148
Query: 431 LLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYLHASD--VMHRDVKS 488
LLVYE++ G++ +HL S L W++RL +AL A+ LA+LH ++ V++RD K+
Sbjct: 149 -LLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLKVALGAAKGLAFLHNAETSVIYRDFKT 207
Query: 489 NNILLDEKFHVKVADFGLSRLFPN-DVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSF 547
+NILLD +++ K++DFGL++ P D +HVST GT GY PEY LT KSDVYS+
Sbjct: 208 SNILLDSEYNAKLSDFGLAKDGPTGDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSY 267
Query: 548 GVVLVELISSLQAVDITRHRNDVNLANMAVNKI-QSQELYDLVDPNLGYEKDNSVKRMTT 606
GVVL+E++S +AVD R + L A + ++L+ ++D L +D
Sbjct: 268 GVVLLEVLSGRRAVDKNRPPGEQKLVEWARPLLANKRKLFRVIDNRL---QDQYSMEEAC 324
Query: 607 AVAELAFRCLQQQRDLRPSMDEIVEVLRAIKS 638
VA LA RCL + LRP+M+E+V L I++
Sbjct: 325 KVATLALRCLTFEIKLRPNMNEVVSHLEHIQT 356
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 187 bits (475), Expect = 8e-47
Identities = 114/289 (39%), Positives = 168/289 (57%), Gaps = 9/289 (3%)
Query: 352 EELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILAR 411
+++ ++T++F+ + +G GGFG VYK L DG VA+KR + +F EVE L+R
Sbjct: 734 DDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSR 793
Query: 412 LRHKNLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETA 470
+H NLV L G C K+ + LL+Y Y+ NG++ LH L W RL IA A
Sbjct: 794 AQHPNLVHLLGYCNYKNDK--LLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAA 851
Query: 471 EALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAPQGTPGY 527
E LAYLH S ++HRD+KS+NILL + F +ADFGL+RL THV+T GT GY
Sbjct: 852 EGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTDLVGTLGY 911
Query: 528 VDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYD 587
+ PEY Q T K DVYSFGVVL+EL++ + +D+ + R +L + + + +
Sbjct: 912 IPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESE 971
Query: 588 LVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAI 636
+ DP + Y+KD++ + + V E+A RCL + RP+ ++V L I
Sbjct: 972 IFDPFI-YDKDHAEEML--LVLEIACRCLGENPKTRPTTQQLVSWLENI 1017
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 180 bits (457), Expect = 1e-44
Identities = 117/314 (37%), Positives = 179/314 (56%), Gaps = 20/314 (6%)
Query: 349 FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKD----------GRVVAVKRHYESNFKR 398
F+ EL+ AT NF +GEGGFG V+KG + + G V+AVKR + F+
Sbjct: 56 FSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQG 115
Query: 399 VAQFMNEVEILARLRHKNLVTLYG-CTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLL 457
+++ E+ L +L H NLV L G C + R LLVYE+++ G++ +HL + L
Sbjct: 116 HREWLAEINYLGQLDHPNLVKLIGYCLEEEHR--LLVYEFMTRGSLENHLFRRGTFYQPL 173
Query: 458 PWSVRLDIALETAEALAYLHASD--VMHRDVKSNNILLDEKFHVKVADFGLSRLFP-NDV 514
W+ R+ +AL A LA+LH + V++RD K++NILLD ++ K++DFGL+R P D
Sbjct: 174 SWNTRVRMALGAARGLAFLHNAQPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDN 233
Query: 515 THVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLAN 574
+HVST GT GY PEY L+ KSDVYSFGVVL+EL+S +A+D + + NL +
Sbjct: 234 SHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHNLVD 293
Query: 575 MAVNKI-QSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
A + + L ++DP L + S+ R +A LA C+ RP+M+EIV+ +
Sbjct: 294 WARPYLTNKRRLLRVMDPRL--QGQYSLTR-ALKIAVLALDCISIDAKSRPTMNEIVKTM 350
Query: 634 RAIKSDEPETQESK 647
+ + ++E +
Sbjct: 351 EELHIQKEASKEQQ 364
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 179 bits (454), Expect = 2e-44
Identities = 120/325 (36%), Positives = 183/325 (55%), Gaps = 35/325 (10%)
Query: 349 FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEI 408
F Y E+ TNNF + +G+GGFG VY G + +G VAVK E + + +F EV++
Sbjct: 564 FKYSEVVNITNNFE--RVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFRAEVDL 620
Query: 409 LARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALE 468
L R+ H NL +L G ++ + ++L+YEY++N + D+L G RS +L W RL I+L+
Sbjct: 621 LMRVHHTNLTSLVGYCNEINH-MVLIYEYMANENLGDYLAGKRSF--ILSWEERLKISLD 677
Query: 469 TAEALAYLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVT-HVSTAPQGT 524
A+ L YLH ++HRDVK NILL+EK K+ADFGLSR F + + +ST G+
Sbjct: 678 AAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGS 737
Query: 525 PGYVDPEYYQCYQLTDKSDVYSFGVVLVELI--------SSLQAVDITRHRNDVNLANMA 576
GY+DPEYY Q+ +KSDVYS GVVL+E+I S + V I+ H + LAN
Sbjct: 738 IGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEKVHISDHVRSI-LANGD 796
Query: 577 VNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIV----EV 632
+ I Q L + D ++ ++E+A C + RP+M ++V ++
Sbjct: 797 IRGIVDQRLRERYDVGSAWK-----------MSEIALACTEHTSAQRPTMSQVVMELKQI 845
Query: 633 LRAIKSDEPETQES-KVLDVVVRTD 656
+ I +D+ +S K+L V + T+
Sbjct: 846 VYGIVTDQENYDDSTKMLTVNLDTE 870
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 179 bits (453), Expect = 3e-44
Identities = 114/345 (33%), Positives = 187/345 (54%), Gaps = 23/345 (6%)
Query: 307 STTSGTNTGTLTSTTNSSQSIPSYPSSKTSTMPKSFYFGVQVFTYEELEEATNNFHTSKE 366
S+T+G T + + I + S + + +S ++V+ + +L+ AT NF
Sbjct: 34 SSTTGATTNSSVGQQSQFSDISTGIISDSGKLLESP--NLKVYNFLDLKTATKNFKPDSM 91
Query: 367 LGEGGFGTVYKG----------DLKDGRVVAVKRHYESNFKRVAQFMNEVEILARLRHKN 416
LG+GGFG VY+G + G +VA+KR + + A++ +EV L L H+N
Sbjct: 92 LGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGFAEWRSEVNFLGMLSHRN 151
Query: 417 LVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALAYL 476
LV L G + +ELLLVYE++ G++ HL PW +R+ I + A LA+L
Sbjct: 152 LVKLLGYC-REDKELLLVYEFMPKGSLESHLFRRNDP---FPWDLRIKIVIGAARGLAFL 207
Query: 477 HA--SDVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDV-THVSTAPQGTPGYVDPEYY 533
H+ +V++RD K++NILLD + K++DFGL++L P D +HV+T GT GY PEY
Sbjct: 208 HSLQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTRIMGTYGYAAPEYM 267
Query: 534 QCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQ-ELYDLVDPN 592
L KSDV++FGVVL+E+++ L A + R R +L + ++ ++ + ++D
Sbjct: 268 ATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLVDWLRPELSNKHRVKQIMDKG 327
Query: 593 LGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIK 637
+ K ++ T +A + C++ RP M E+VEVL I+
Sbjct: 328 I---KGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVLEHIQ 369
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 174 bits (442), Expect = 5e-43
Identities = 115/324 (35%), Positives = 167/324 (51%), Gaps = 24/324 (7%)
Query: 334 KTSTMPKSFYFGVQVFTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKR--- 390
K+ST+ S + + E E + +G G G VYK +L+ G VVAVK+
Sbjct: 657 KSSTLAASKWRSFHKLHFSE-HEIADCLDEKNVIGFGSSGKVYKVELRGGEVVAVKKLNK 715
Query: 391 -------HYESNFKRVAQFMNEVEILARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTV 443
Y S+ F EVE L +RHK++V L+ C S +LL VYEY+ NG++
Sbjct: 716 SVKGGDDEYSSDSLNRDVFAAEVETLGTIRHKSIVRLWCCCSSGDCKLL-VYEYMPNGSL 774
Query: 444 ADHLHGDRSSSCLLPWSVRLDIALETAEALAYLH---ASDVMHRDVKSNNILLDEKFHVK 500
AD LHGDR +L W RL IAL+ AE L+YLH ++HRDVKS+NILLD + K
Sbjct: 775 ADVLHGDRKGGVVLGWPERLRIALDAAEGLSYLHHDCVPPIVHRDVKSSNILLDSDYGAK 834
Query: 501 VADFGLSR---LFPNDVTHVSTAPQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISS 557
VADFG+++ + + + G+ GY+ PEY ++ +KSD+YSFGVVL+EL++
Sbjct: 835 VADFGIAKVGQMSGSKTPEAMSGIAGSCGYIAPEYVYTLRVNEKSDIYSFGVVLLELVTG 894
Query: 558 LQAVDITRHRNDVNLANMAVNKIQSQELYDLVDPNLGYEKDNSVKRMTTAVAELAFRCLQ 617
Q D D ++A + L ++DP L D K + V + C
Sbjct: 895 KQPTD--SELGDKDMAKWVCTALDKCGLEPVIDPKL----DLKFKEEISKVIHIGLLCTS 948
Query: 618 QQRDLRPSMDEIVEVLRAIKSDEP 641
RPSM ++V +L+ + P
Sbjct: 949 PLPLNRPSMRKVVIMLQEVSGAVP 972
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 174 bits (440), Expect = 9e-43
Identities = 101/299 (33%), Positives = 171/299 (56%), Gaps = 10/299 (3%)
Query: 352 EELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEILAR 411
E + +AT NF + +LG+GGFG VYKG L DG+ +AVKR +++ + +FMNEV ++AR
Sbjct: 519 ETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIAR 578
Query: 412 LRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAE 471
L+H NLV + GC + E +L+YEY+ N ++ +L G ++ L W+ R DI A
Sbjct: 579 LQHINLVQVLGCCIE-GDEKMLIYEYLENLSLDSYLFG-KTRRSKLNWNERFDITNGVAR 636
Query: 472 ALAYLHASD---VMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHVSTAP-QGTPGY 527
L YLH ++HRD+K +NILLD+ K++DFG++R+F D T +T GT GY
Sbjct: 637 GLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGY 696
Query: 528 VDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYD 587
+ PEY ++KSDV+SFGV+++E++S + + +L + ++ + +
Sbjct: 697 MSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRALE 756
Query: 588 LVDP----NLGYEKDNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVLRAIKSDEPE 642
+VDP +L + + ++ C+Q+ + RP+M +V + + ++ P+
Sbjct: 757 IVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVWMFGSEATEIPQ 815
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 164 bits (415), Expect = 7e-40
Identities = 112/299 (37%), Positives = 168/299 (55%), Gaps = 24/299 (8%)
Query: 349 FTYEELEEATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFMNEVEI 408
++Y EL +AT F ELG G GTVYKG L+D R VAVK+ E+ + F E+ +
Sbjct: 524 YSYRELVKATRKFKV--ELGRGESGTVYKGVLEDDRHVAVKK-LENVRQGKEVFQAELSV 580
Query: 409 LARLRHKNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALE 468
+ R+ H NLV ++G S+ S LL V EY+ NG++A+ L + + LL W R +IAL
Sbjct: 581 IGRINHMNLVRIWGFCSEGSHRLL-VSEYVENGSLANILFSE-GGNILLDWEGRFNIALG 638
Query: 469 TAEALAYLHASD---VMHRDVKSNNILLDEKFHVKVADFGLSRLF-----PNDVTHVSTA 520
A+ LAYLH V+H DVK NILLD+ F K+ DFGL +L +V+HV
Sbjct: 639 VAKGLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHV--- 695
Query: 521 PQGTPGYVDPEYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVN-----LANM 575
+GT GY+ PE+ +T K DVYS+GVVL+EL++ + ++ ++V+ L M
Sbjct: 696 -RGTLGYIAPEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRM 754
Query: 576 AVNKIQSQELYDLVDPNLGYEKDNSVKRM-TTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
K++ +E +D L + + V + + +LA CL++ R RP+M+ V+ L
Sbjct: 755 LSAKLEGEE-QSWIDGYLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAVQTL 812
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 157 bits (397), Expect = 9e-38
Identities = 103/284 (36%), Positives = 159/284 (55%), Gaps = 8/284 (2%)
Query: 356 EATNNFHTSKELGEGGFGTVYKGDLKDGRVVAVKRHYESNFKRVAQFM-NEVEILARLRH 414
EAT N + +G+G GT+YK L +V AVK+ + K + M E+E + ++RH
Sbjct: 811 EATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKVRH 870
Query: 415 KNLVTLYGCTSKHSRELLLVYEYISNGTVADHLHGDRSSSCLLPWSVRLDIALETAEALA 474
+NL+ L + L+L Y Y+ NG++ D LH + + L WS R +IA+ TA LA
Sbjct: 871 RNLIKLEEFWLRKEYGLIL-YTYMENGSLHDILH-ETNPPKPLDWSTRHNIAVGTAHGLA 928
Query: 475 YLHAS---DVMHRDVKSNNILLDEKFHVKVADFGLSRLFPNDVTHV-STAPQGTPGYVDP 530
YLH ++HRD+K NILLD ++DFG+++L T + S QGT GY+ P
Sbjct: 929 YLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAP 988
Query: 531 EYYQCYQLTDKSDVYSFGVVLVELISSLQAVDITRHRNDVNLANMAVNKIQSQELYDLVD 590
E + +SDVYS+GVVL+ELI+ +A+D + + + + Q+ E+ +VD
Sbjct: 989 ENAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNGETDIVGWVRSVWTQTGEIQKIVD 1048
Query: 591 PNLGYEK-DNSVKRMTTAVAELAFRCLQQQRDLRPSMDEIVEVL 633
P+L E D+SV T LA RC +++ D RP+M ++V+ L
Sbjct: 1049 PSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 84,610,013
Number of Sequences: 164201
Number of extensions: 3813881
Number of successful extensions: 16578
Number of sequences better than 10.0: 1690
Number of HSP's better than 10.0 without gapping: 1340
Number of HSP's successfully gapped in prelim test: 350
Number of HSP's that attempted gapping in prelim test: 12357
Number of HSP's gapped (non-prelim): 2143
length of query: 687
length of database: 59,974,054
effective HSP length: 117
effective length of query: 570
effective length of database: 40,762,537
effective search space: 23234646090
effective search space used: 23234646090
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147875.4


