

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147875.1 + phase: 0
(371 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
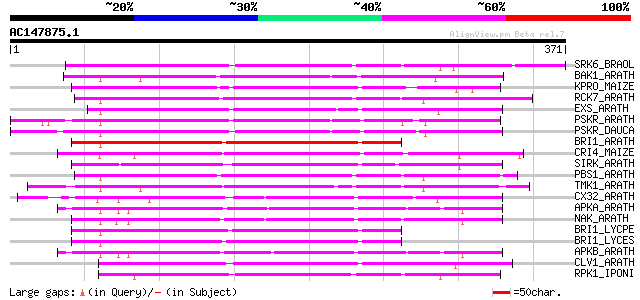
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 191 3e-48
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 185 1e-46
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 181 3e-45
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 172 9e-43
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 171 4e-42
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 169 8e-42
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 167 3e-41
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 164 3e-40
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 161 2e-39
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 161 3e-39
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 159 8e-39
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 157 3e-38
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 156 9e-38
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 155 1e-37
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 154 3e-37
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 154 3e-37
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 154 5e-37
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 152 1e-36
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 152 2e-36
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 150 4e-36
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 191 bits (484), Expect = 3e-48
Identities = 118/345 (34%), Positives = 191/345 (55%), Gaps = 18/345 (5%)
Query: 38 LSMPRRYSYAEVKMITNYFR-EKLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFI 95
L +P VK N+ KLGQGG+G+VYK L + +++AVK +S+T G +EF+
Sbjct: 511 LELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFM 570
Query: 96 NEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTL 155
NEV I+R H+N+V +LG C E +++ LIYE++ SLD +++ + + +WN
Sbjct: 571 NEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKL---NWNER 627
Query: 156 FRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSI 215
F I G+A+GL YLHQ RI+H D+K NILLD++ PKISDFG+A+I +R ++ +
Sbjct: 628 FDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANT 687
Query: 216 LGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW 275
+ GT GYM+PE +G S +SDV+S+G+++LE++ G+KN + E +
Sbjct: 688 MKVVGTYGYMSPEY--AMYGIFSEKSDVFSFGVIVLEIVSGKKNRGF-YNLDYENDLLSY 744
Query: 276 IYKDLEQGNTL-----LNCSTISEE----ENDMIRKITLVSLWCIQTKPSDRPPMNKVIE 326
++ ++G L + ++S + + + K + L C+Q RP M+ V+
Sbjct: 745 VWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVVW 804
Query: 327 MLQGPLSSVSYPPKPVLYSPERPELQVSDMSSSDLYETNSVTVSK 371
M + + PKP Y R ++ SS E S TV++
Sbjct: 805 MFGSEATEIP-QPKPPGYCVRRSPYELDPSSSWQCDENESWTVNQ 848
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 185 bits (470), Expect = 1e-46
Identities = 109/301 (36%), Positives = 174/301 (57%), Gaps = 12/301 (3%)
Query: 37 NLSMPRRYSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISE--TKGNGE 92
+L +R+S E+++ ++ F K LG+GG+G VYK L + VAVK + E T+G
Sbjct: 270 HLGQLKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGEL 329
Query: 93 EFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDW 152
+F EV IS H N++ L G+C +R L+Y +M GS+ + E P + DW
Sbjct: 330 QFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCL--RERPESQPPLDW 387
Query: 153 NTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSI 212
RIA+G A+GL YLH C +I+H D+K NILLDE+F + DFGLAK+ KD+
Sbjct: 388 PKRQRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTH 447
Query: 213 VSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCT-SEMY 271
V+ RGTIG++APE S G S ++DV+ YG+++LE+I G++ +D ++
Sbjct: 448 VT-TAVRGTIGHIAPEYLST--GKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVM 504
Query: 272 FPDWIYKDLEQG--NTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
DW+ L++ L++ +++ + ++ V+L C Q+ P +RP M++V+ ML+
Sbjct: 505 LLDWVKGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLE 564
Query: 330 G 330
G
Sbjct: 565 G 565
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 181 bits (459), Expect = 3e-45
Identities = 109/303 (35%), Positives = 167/303 (54%), Gaps = 28/303 (9%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNGEEFINEVASI 101
RRYSY E+ T F+ +LG+G G VYK L + R VAVK + + E F E++ I
Sbjct: 522 RRYSYRELVKATRKFKVELGRGESGTVYKGVLEDDRHVAVKKLENVRQGKEVFQAELSVI 581
Query: 102 SRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIG 161
R +HMN+V + G+C E + R L+ E++ GSL ++ SE N + DW F IA+G
Sbjct: 582 GRINHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILF-SEGGNIL--LDWEGRFNIALG 638
Query: 162 IAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGT 221
+AKGL YLH C ++H D+KP+NILLD+ F PKI+DFGL K+ R S ++ RGT
Sbjct: 639 VAKGLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGT 698
Query: 222 IGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLE 281
+GY+APE S ++ + DVYSYG+++LE++ G + + G D ++ L
Sbjct: 699 LGYIAPEWVSSL--PITAKVDVYSYGVVLLELLTGTRVSELVGG-------TDEVHSMLR 749
Query: 282 QGNTLLNCSTISEEEN--------------DMIRKITLVSL--WCIQTKPSDRPPMNKVI 325
+ +L+ EE++ + ++ TL+ L C++ S RP M +
Sbjct: 750 KLVRMLSAKLEGEEQSWIDGYLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAV 809
Query: 326 EML 328
+ L
Sbjct: 810 QTL 812
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 172 bits (437), Expect = 9e-43
Identities = 106/313 (33%), Positives = 170/313 (53%), Gaps = 12/313 (3%)
Query: 44 YSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQV-AVKVISETKGNG-EEFINEVA 99
+++ E+ T FR LG+GG+G V+K ++ QV A+K + G EF+ EV
Sbjct: 91 FTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEVL 150
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIA 159
++S H N+V L+G+C E ++R L+YE+MP+GSL+ ++ P+ DWNT +IA
Sbjct: 151 TLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHV--LPSGKKPLDWNTRMKIA 208
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
G A+GLEYLH + +++ D+K NILL ED+ PK+SDFGLAK+ D
Sbjct: 209 AGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVSTRVM 268
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW---I 276
GT GY AP+ G ++++SD+YS+G+++LE+I GRK D + W +
Sbjct: 269 GTYGYCAPDY--AMTGQLTFKSDIYSFGVVLLELITGRKAID-NTKTRKDQNLVGWARPL 325
Query: 277 YKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVS 336
+KD +++ + + + +S C+Q +P+ RP ++ V+ L SS
Sbjct: 326 FKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALNFLASSKY 385
Query: 337 YPPKPVLYSPERP 349
P P S + P
Sbjct: 386 DPNSPSSSSGKNP 398
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 171 bits (432), Expect = 4e-42
Identities = 100/282 (35%), Positives = 161/282 (56%), Gaps = 10/282 (3%)
Query: 53 TNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVASISRTSHMNI 109
T++F +K +G GG+G VYKA L + VAVK +SE K G EF+ E+ ++ + H N+
Sbjct: 914 TDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVKHPNL 973
Query: 110 VSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYL 169
VSLLGYC ++ L+YE+M GSLD ++ + DW+ +IA+G A+GL +L
Sbjct: 974 VSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEV--LDWSKRLKIAVGAARGLAFL 1031
Query: 170 HQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEI 229
H G I+H DIK NILLD DF PK++DFGLA++ +S VS + A GT GY+ PE
Sbjct: 1032 HHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIA-GTFGYIPPEY 1090
Query: 230 FSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGNT--LL 287
A + + DVYS+G+++LE++ G++ + W + + QG ++
Sbjct: 1091 GQSA--RATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAIQKINQGKAVDVI 1148
Query: 288 NCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
+ +S + ++ +++ C+ P+ RP M V++ L+
Sbjct: 1149 DPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 169 bits (429), Expect = 8e-42
Identities = 113/341 (33%), Positives = 188/341 (54%), Gaps = 25/341 (7%)
Query: 1 MIILRQQQQSSLIPQTILRE----RTEL--VDNNVEVFMRSHNLSMPRRYSYAEVKMITN 54
+I+LR +++S + I R EL + + + V +S++ + SY ++ TN
Sbjct: 677 LIVLRARRRSGEVDPEIEESESMNRKELGEIGSKLVVLFQSND----KELSYDDLLDSTN 732
Query: 55 YFREK--LGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVASISRTSHMNIVS 111
F + +G GG+G+VYKA+L + ++VA+K +S G E EF EV ++SR H N+V
Sbjct: 733 SFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRAQHPNLVL 792
Query: 112 LLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQ 171
L G+C+ +N R LIY +M GSLD ++++ A+ W T RIA G AKGL YLH+
Sbjct: 793 LRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPAL--LKWKTRLRIAQGAAKGLLYLHE 850
Query: 172 GCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFS 231
GC ILH DIK NILLDE+F ++DFGLA++ ++ VS GT+GY+ PE
Sbjct: 851 GCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVS-TDLVGTLGYIPPEYGQ 909
Query: 232 RAFGGVSYRSDVYSYGMLILEMIGGRKNYD--TGGSCTSEMYFPDWI--YKDLEQGNTLL 287
+ +Y+ DVYS+G+++LE++ ++ D C + W+ K + + +
Sbjct: 910 ASV--ATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLI---SWVVKMKHESRASEVF 964
Query: 288 NCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEML 328
+ S+E + + ++ ++ C+ P RP +++ L
Sbjct: 965 DPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 167 bits (424), Expect = 3e-41
Identities = 111/336 (33%), Positives = 181/336 (53%), Gaps = 17/336 (5%)
Query: 1 MIILRQQQQSSLIPQTIL-RERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREK 59
+IILR + + P+ + EL +V +F HN S ++ T+ F +
Sbjct: 690 LIILRTTSRGEVDPEKKADADEIELGSRSVVLF---HNKDSNNELSLDDILKSTSSFNQA 746
Query: 60 --LGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEVASISRTSHMNIVSLLGYC 116
+G GG+G+VYKA+L + +VA+K +S G + EF EV ++SR H N+V LLGYC
Sbjct: 747 NIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSRAQHPNLVHLLGYC 806
Query: 117 YEENKRALIYEFMPKGSLDKFIY-KSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSS 175
+N + LIY +M GSLD +++ K + P ++ DW T RIA G A+GL YLHQ C
Sbjct: 807 NYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSL---DWKTRLRIARGAAEGLAYLHQSCEP 863
Query: 176 RILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFG 235
ILH DIK NILL + F ++DFGLA++ D+ V+ GT+GY+ PE +
Sbjct: 864 HILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVT-TDLVGTLGYIPPEYGQASV- 921
Query: 236 GVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWI--YKDLEQGNTLLNCSTIS 293
+Y+ DVYS+G+++LE++ GR+ D S W+ K ++ + + +
Sbjct: 922 -ATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLI-SWVLQMKTEKRESEIFDPFIYD 979
Query: 294 EEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
++ + + + ++ C+ P RP +++ L+
Sbjct: 980 KDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 164 bits (415), Expect = 3e-40
Identities = 87/224 (38%), Positives = 138/224 (60%), Gaps = 7/224 (3%)
Query: 42 RRYSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEV 98
R+ ++A++ TN F +G GG+G VYKA L + VA+K + G G+ EF+ E+
Sbjct: 869 RKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEM 928
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
+I + H N+V LLGYC ++R L+YEFM GSL+ ++ + A +W+T +I
Sbjct: 929 ETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPK--KAGVKLNWSTRRKI 986
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGA 218
AIG A+GL +LH CS I+H D+K N+LLDE+ ++SDFG+A++ D+ +S+
Sbjct: 987 AIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTL 1046
Query: 219 RGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDT 262
GT GY+ PE + S + DVYSYG+++LE++ G++ D+
Sbjct: 1047 AGTPGYVPPEYYQSF--RCSTKGDVYSYGVVLLELLTGKRPTDS 1088
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 161 bits (408), Expect = 2e-39
Identities = 101/321 (31%), Positives = 174/321 (53%), Gaps = 16/321 (4%)
Query: 33 MRSHNLSMPRRYSYAEVKMITNYFRE--KLGQGGYGVVYKASLYNSRQVAVK---VISET 87
M + + +SY E++ T F E ++G+G + V+K L + VAVK S+
Sbjct: 482 MEDLKIRRAQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDV 541
Query: 88 KGNGEEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAI 147
K + +EF NE+ +SR +H ++++LLGYC + ++R L+YEFM GSL + ++ + PN
Sbjct: 542 KKSSKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKD-PNLK 600
Query: 148 CDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQ 207
+W IA+ A+G+EYLH ++H DIK NIL+DED +++DFGL+ +
Sbjct: 601 KRLNWARRVTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGP 660
Query: 208 RKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCT 267
GT+GY+ PE + + ++ +SDVYS+G+++LE++ GRK D
Sbjct: 661 ADSGTPLSELPAGTLGYLDPEYYRLHY--LTTKSDVYSFGVVLLEILSGRKAIDMQ---F 715
Query: 268 SEMYFPDWIYKDLEQGNTLLNCSTISEEENDM--IRKITLVSLWCIQTKPSDRPPMNKVI 325
E +W ++ G+ + +D+ ++KI V+ C++ + DRP M+KV
Sbjct: 716 EEGNIVEWAVPLIKAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVT 775
Query: 326 EMLQGPLSSVSYPP---KPVL 343
L+ L+ + P +P+L
Sbjct: 776 TALEHALALLMGSPCIEQPIL 796
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 161 bits (407), Expect = 3e-39
Identities = 105/292 (35%), Positives = 159/292 (53%), Gaps = 14/292 (4%)
Query: 42 RRYSYAEVKMITNYFREKLGQGGYGVVYKASLYNSRQVAVKVISETKGNG-EEFINEVAS 100
R + Y+EV ITN F +G+GG+G VY + N QVAVKV+SE G +EF EV
Sbjct: 562 RYFKYSEVVNITNNFERVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFRAEVDL 620
Query: 101 ISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY-KSEFPNAICDFDWNTLFRIA 159
+ R H N+ SL+GYC E N LIYE+M +L ++ K F W +I+
Sbjct: 621 LMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSF-----ILSWEERLKIS 675
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
+ A+GLEYLH GC I+H D+KP NILL+E K++DFGL++ + S
Sbjct: 676 LDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVA 735
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKD 279
G+IGY+ PE +S ++ +SDVYS G+++LE+I G+ S T +++ D +
Sbjct: 736 GSIGYLDPEYYSTR--QMNEKSDVYSLGVVLLEVITGQP--AIASSKTEKVHISDHVRSI 791
Query: 280 LEQGNTLLNCSTISEEENDM--IRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
L G+ E D+ K++ ++L C + + RP M++V+ L+
Sbjct: 792 LANGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELK 843
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 159 bits (403), Expect = 8e-39
Identities = 109/303 (35%), Positives = 161/303 (52%), Gaps = 13/303 (4%)
Query: 44 YSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQV-AVKVISETKGNGE-EFINEVA 99
+++ E+ T F LG+GG+G VYK L ++ QV AVK + G EF+ EV
Sbjct: 74 FAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVL 133
Query: 100 SISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIA 159
+S H N+V+L+GYC + ++R L+YEFMP GSL+ ++ + P DWN +IA
Sbjct: 134 MLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLH--DLPPDKEALDWNMRMKIA 191
Query: 160 IGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGAR 219
G AKGLE+LH + +++ D K NILLDE F PK+SDFGLAK+ D
Sbjct: 192 AGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVM 251
Query: 220 GTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDW---I 276
GT GY APE G ++ +SDVYS+G++ LE+I GRK D+ E W +
Sbjct: 252 GTYGYCAPEY--AMTGQLTVKSDVYSFGVVFLELITGRKAIDS-EMPHGEQNLVAWARPL 308
Query: 277 YKDLEQGNTLLNCSTISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVS 336
+ D + L + + + V+ CIQ + + RP + V+ L L++ +
Sbjct: 309 FNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL-SYLANQA 367
Query: 337 YPP 339
Y P
Sbjct: 368 YDP 370
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 157 bits (398), Expect = 3e-38
Identities = 106/346 (30%), Positives = 179/346 (51%), Gaps = 26/346 (7%)
Query: 13 IPQTILRERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFREK--LGQGGYGVVYK 70
I T T V +N+++ + L S ++ +TN F LG GG+GVVYK
Sbjct: 550 ISDTYTLPGTSEVGDNIQMVEAGNML-----ISIQVLRSVTNNFSSDNILGSGGFGVVYK 604
Query: 71 ASLYNSRQVAVKVISE--TKGNG-EEFINEVASISRTSHMNIVSLLGYCYEENKRALIYE 127
L++ ++AVK + G G EF +E+A +++ H ++V+LLGYC + N++ L+YE
Sbjct: 605 GELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHRHLVTLLGYCLDGNEKLLVYE 664
Query: 128 FMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNI 187
+MP+G+L + +++ + W +A+ +A+G+EYLH +H D+KP NI
Sbjct: 665 YMPQGTLSRHLFEWS-EEGLKPLLWKQRLTLALDVARGVEYLHGLAHQSFIHRDLKPSNI 723
Query: 188 LLDEDFCPKISDFGLAKIC-QRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSY 246
LL +D K++DFGL ++ + K SI + + GT GY+APE G V+ + DVYS+
Sbjct: 724 LLGDDMRAKVADFGLVRLAPEGKGSIETRIA--GTFGYLAPEY--AVTGRVTTKVDVYSF 779
Query: 247 GMLILEMIGGRKNYDTGGSCTSEMYFPDW-----IYKDLEQGNTLLNCSTISEEENDMIR 301
G++++E+I GRK+ D ++ W I K+ + + EE +
Sbjct: 780 GVILMELITGRKSLDE-SQPEESIHLVSWFKRMYINKEASFKKAIDTTIDLDEETLASVH 838
Query: 302 KITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVSYPPKPVLYSPE 347
+ ++ C +P RP M + + LSS+ KP +PE
Sbjct: 839 TVAELAGHCCAREPYQRPDMGHAVNI----LSSLVELWKPSDQNPE 880
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 156 bits (394), Expect = 9e-38
Identities = 107/340 (31%), Positives = 179/340 (52%), Gaps = 36/340 (10%)
Query: 6 QQQQSSLIPQTILRERTELVDNNVEVFMRSHNLSMPRRYSYAEVKMITNYFR--EKLGQG 63
QQ Q S I I+ + +L++ S NL + Y++ ++K T F+ LGQG
Sbjct: 47 QQSQFSDISTGIISDSGKLLE--------SPNLKV---YNFLDLKTATKNFKPDSMLGQG 95
Query: 64 GYGVVYKA-----SLYNSRQVAVKVISETKGNGE------EFINEVASISRTSHMNIVSL 112
G+G VY+ +L SR + +++ + N E E+ +EV + SH N+V L
Sbjct: 96 GFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKL 155
Query: 113 LGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQG 172
LGYC E+ + L+YEFMPKGSL+ +++ P F W+ +I IG A+GL +LH
Sbjct: 156 LGYCREDKELLLVYEFMPKGSLESHLFRRNDP-----FPWDLRIKIVIGAARGLAFLH-S 209
Query: 173 CSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSR 232
+++ D K NILLD ++ K+SDFGLAK+ + GT GY APE
Sbjct: 210 LQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTRIMGTYGYAAPEYM-- 267
Query: 233 AFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGN---TLLNC 289
A G + +SDV+++G+++LE++ G ++T E DW+ +L + +++
Sbjct: 268 ATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQES-LVDWLRPELSNKHRVKQIMDK 326
Query: 290 STISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQ 329
+ + ++ ++L CI+ P +RP M +V+E+L+
Sbjct: 327 GIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVLE 366
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 155 bits (393), Expect = 1e-37
Identities = 107/315 (33%), Positives = 167/315 (52%), Gaps = 29/315 (9%)
Query: 33 MRSHNLSMPRRYSYAEVKMITNYFREK--LGQGGYGVVYKA-----SLYNSRQ-----VA 80
++S NL + +S+AE+K T FR LG+GG+G V+K SL SR +A
Sbjct: 48 LQSPNL---KSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIA 104
Query: 81 VKVISETKGNG-EEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY 139
VK +++ G +E++ EV + + SH ++V L+GYC E+ R L+YEFMP+GSL+ ++
Sbjct: 105 VKKLNQDGWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF 164
Query: 140 KSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISD 199
+ W ++A+G AKGL +LH +R+++ D K NILLD ++ K+SD
Sbjct: 165 RRGL--YFQPLSWKLRLKVALGAAKGLAFLHSS-ETRVIYRDFKTSNILLDSEYNAKLSD 221
Query: 200 FGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKN 259
FGLAK D GT GY APE A G ++ +SDVYS+G+++LE++ GR+
Sbjct: 222 FGLAKDGPIGDKSHVSTRVMGTHGYAAPEYL--ATGHLTTKSDVYSFGVVLLELLSGRRA 279
Query: 260 YDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIR-----KITLVSLWCIQTK 314
D + E +W L + I D K+ +SL C+ T+
Sbjct: 280 VDKNRP-SGERNLVEWAKPYLVNKRKIFR--VIDNRLQDQYSMEEACKVATLSLRCLTTE 336
Query: 315 PSDRPPMNKVIEMLQ 329
RP M++V+ L+
Sbjct: 337 IKLRPNMSEVVSHLE 351
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 154 bits (389), Expect = 3e-37
Identities = 102/304 (33%), Positives = 163/304 (53%), Gaps = 22/304 (7%)
Query: 42 RRYSYAEVKMITNYFREK--LGQGGYGVVYK-----ASLYNSRQ-----VAVKVISETKG 89
+ +S +E+K T FR +G+GG+G V+K +SL S+ +AVK +++
Sbjct: 54 KNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGF 113
Query: 90 NGE-EFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAIC 148
G E++ E+ + + H N+V L+GYC EE R L+YEFM +GSL+ +++
Sbjct: 114 QGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRR--GTFYQ 171
Query: 149 DFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQR 208
WNT R+A+G A+GL +LH ++++ D K NILLD ++ K+SDFGLA+
Sbjct: 172 PLSWNTRVRMALGAARGLAFLH-NAQPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPM 230
Query: 209 KDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDTGGSCTS 268
D+ GT GY APE A G +S +SDVYS+G+++LE++ GR+ D
Sbjct: 231 GDNSHVSTRVMGTQGYAAPEYL--ATGHLSVKSDVYSFGVVLLELLSGRRAIDK-NQPVG 287
Query: 269 EMYFPDWIYKDLEQGNTLLN-CSTISEEENDMIR--KITLVSLWCIQTKPSDRPPMNKVI 325
E DW L LL + + + R KI +++L CI RP MN+++
Sbjct: 288 EHNLVDWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIV 347
Query: 326 EMLQ 329
+ ++
Sbjct: 348 KTME 351
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 154 bits (389), Expect = 3e-37
Identities = 83/224 (37%), Positives = 132/224 (58%), Gaps = 7/224 (3%)
Query: 42 RRYSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEV 98
R+ ++A++ TN F +G GG+G VYKA L + VA+K + G G+ EF E+
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEM 933
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
+I + H N+V LLGYC +R L+YE+M GSL+ ++ + +W +I
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGI--KLNWPARRKI 991
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGA 218
AIG A+GL +LH C I+H D+K N+LLDE+ ++SDFG+A++ D+ +S+
Sbjct: 992 AIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTL 1051
Query: 219 RGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDT 262
GT GY+ PE + S + DVYSYG+++LE++ G++ D+
Sbjct: 1052 AGTPGYVPPEYYQSF--RCSTKGDVYSYGVVLLELLTGKQPTDS 1093
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 154 bits (388), Expect = 5e-37
Identities = 83/224 (37%), Positives = 132/224 (58%), Gaps = 7/224 (3%)
Query: 42 RRYSYAEVKMITNYFREK--LGQGGYGVVYKASLYNSRQVAVKVISETKGNGE-EFINEV 98
R+ ++A++ TN F +G GG+G VYKA L + VA+K + G G+ EF E+
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEM 933
Query: 99 ASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRI 158
+I + H N+V LLGYC +R L+YE+M GSL+ ++ + +W +I
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRK--KIGIKLNWPARRKI 991
Query: 159 AIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGA 218
AIG A+GL +LH C I+H D+K N+LLDE+ ++SDFG+A++ D+ +S+
Sbjct: 992 AIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTL 1051
Query: 219 RGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKNYDT 262
GT GY+ PE + S + DVYSYG+++LE++ G++ D+
Sbjct: 1052 AGTPGYVPPEYYQSF--RCSTKGDVYSYGVVLLELLTGKQPTDS 1093
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 152 bits (385), Expect = 1e-36
Identities = 106/315 (33%), Positives = 166/315 (52%), Gaps = 29/315 (9%)
Query: 33 MRSHNLSMPRRYSYAEVKMITNYFREK--LGQGGYGVVYKA-----SLYNSRQ-----VA 80
++S NL + +++AE+K T FR LG+GG+G V+K +L S+ +A
Sbjct: 49 LQSPNL---KSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIA 105
Query: 81 VKVISETKGNG-EEFINEVASISRTSHMNIVSLLGYCYEENKRALIYEFMPKGSLDKFIY 139
VK +++ G +E++ EV + + SH N+V L+GYC E+ R L+YEFMP+GSL+ ++
Sbjct: 106 VKKLNQDGWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLF 165
Query: 140 KSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRILHLDIKPQNILLDEDFCPKISD 199
+ + W ++A+G AKGL +LH +S +++ D K NILLD ++ K+SD
Sbjct: 166 RR--GSYFQPLSWTLRLKVALGAAKGLAFLHNAETS-VIYRDFKTSNILLDSEYNAKLSD 222
Query: 200 FGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGVSYRSDVYSYGMLILEMIGGRKN 259
FGLAK D GT GY APE A G ++ +SDVYSYG+++LE++ GR+
Sbjct: 223 FGLAKDGPTGDKSHVSTRIMGTYGYAAPEYL--ATGHLTTKSDVYSYGVVLLEVLSGRRA 280
Query: 260 YDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEENDMIR-----KITLVSLWCIQTK 314
D E +W L L I D K+ ++L C+ +
Sbjct: 281 VDKNRP-PGEQKLVEWARPLLANKRKLFR--VIDNRLQDQYSMEEACKVATLALRCLTFE 337
Query: 315 PSDRPPMNKVIEMLQ 329
RP MN+V+ L+
Sbjct: 338 IKLRPNMNEVVSHLE 352
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 152 bits (383), Expect = 2e-36
Identities = 93/284 (32%), Positives = 151/284 (52%), Gaps = 13/284 (4%)
Query: 60 LGQGGYGVVYKASLYNSRQVAVK-VISETKGNGEE-FINEVASISRTSHMNIVSLLGYCY 117
+G+GG G+VY+ S+ N+ VA+K ++ G + F E+ ++ R H +IV LLGY
Sbjct: 698 IGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRLLGYVA 757
Query: 118 EENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRI 177
++ L+YE+MP GSL + ++ S+ + W T R+A+ AKGL YLH CS I
Sbjct: 758 NKDTNLLLYEYMPNGSLGELLHGSKGGH----LQWETRHRVAVEAAKGLCYLHHDCSPLI 813
Query: 178 LHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGV 237
LH D+K NILLD DF ++DFGLAK + + G+ GY+APE V
Sbjct: 814 LHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTL--KV 871
Query: 238 SYRSDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGNTLLNCSTISEEE- 296
+SDVYS+G+++LE+I G+K G + + +++ Q + I +
Sbjct: 872 DEKSDVYSFGVVLLELIAGKKPVGEFGEGVDIVRWVRNTEEEITQPSDAAIVVAIVDPRL 931
Query: 297 ----NDMIRKITLVSLWCIQTKPSDRPPMNKVIEMLQGPLSSVS 336
+ + +++ C++ + + RP M +V+ ML P SV+
Sbjct: 932 TGYPLTSVIHVFKIAMMCVEEEAAARPTMREVVHMLTNPPKSVA 975
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 150 bits (380), Expect = 4e-36
Identities = 94/279 (33%), Positives = 142/279 (50%), Gaps = 18/279 (6%)
Query: 60 LGQGGYGVVYKASLYNSRQVAVK--VISETKGNGEEFINEVASISRTSHMNIVSLLGYCY 117
+G+G +G +YKA+L + AVK V + K + E+ +I + H N++ L +
Sbjct: 822 IGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKVRHRNLIKLEEFWL 881
Query: 118 EENKRALIYEFMPKGSLDKFIYKSEFPNAICDFDWNTLFRIAIGIAKGLEYLHQGCSSRI 177
+ ++Y +M GSL ++++ P + DW+T IA+G A GL YLH C I
Sbjct: 882 RKEYGLILYTYMENGSLHDILHETNPPKPL---DWSTRHNIAVGTAHGLAYLHFDCDPAI 938
Query: 178 LHLDIKPQNILLDEDFCPKISDFGLAKICQRKDSIVSILGARGTIGYMAPEIFSRAFGGV 237
+H DIKP NILLD D P ISDFG+AK+ + + + +GTIGYMAPE AF V
Sbjct: 939 VHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPE---NAFTTV 995
Query: 238 SYR-SDVYSYGMLILEMIGGRKNYDTGGSCTSEMYFPDWIYKDLEQGNTL-------LNC 289
R SDVYSYG+++LE+I +K D S E W+ Q + L
Sbjct: 996 KSRESDVYSYGVVLLELITRKKALDP--SFNGETDIVGWVRSVWTQTGEIQKIVDPSLLD 1053
Query: 290 STISEEENDMIRKITLVSLWCIQTKPSDRPPMNKVIEML 328
I + + + ++L C + + RP M V++ L
Sbjct: 1054 ELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 43,768,161
Number of Sequences: 164201
Number of extensions: 1899184
Number of successful extensions: 7743
Number of sequences better than 10.0: 1666
Number of HSP's better than 10.0 without gapping: 1028
Number of HSP's successfully gapped in prelim test: 638
Number of HSP's that attempted gapping in prelim test: 4634
Number of HSP's gapped (non-prelim): 1849
length of query: 371
length of database: 59,974,054
effective HSP length: 112
effective length of query: 259
effective length of database: 41,583,542
effective search space: 10770137378
effective search space used: 10770137378
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC147875.1


