

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147712.9 - phase: 0
(512 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
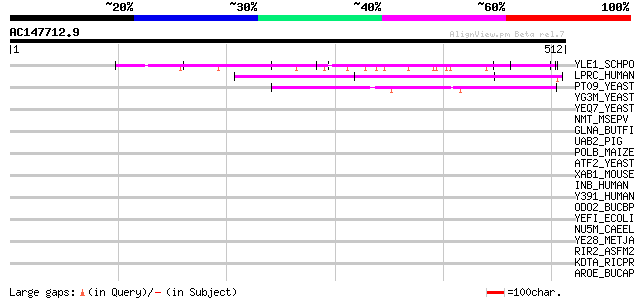
YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I 67 1e-10
LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP13... 65 4e-10
PT09_YEAST (P32522) PET309 protein, mitochondrial precursor 45 6e-04
YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR... 42 0.003
YEQ7_YEAST (P40050) Hypothetical 79.5 kDa protein in PTP3-SER3 i... 39 0.042
NMT_MSEPV (Q9YW20) Putative glycylpeptide N-tetradecanoyltransfe... 38 0.071
GLNA_BUTFI (Q05650) Glutamine synthetase (EC 6.3.1.2) (Glutamate... 35 0.46
UAB2_PIG (P46202) Uteroferrin-associated basic protein 2 precurs... 33 1.3
POLB_MAIZE (P15718) Retrovirus-related Pol polyprotein from tran... 33 1.3
ATF2_YEAST (P53296) Alcohol O-acetyltransferase 2 (EC 2.3.1.84) ... 33 1.3
XAB1_MOUSE (Q8VCE2) XPA-binding protein 1 33 1.8
INB_HUMAN (P01574) Interferon beta precursor (IFN-beta) (Fibrobl... 32 3.0
Y391_HUMAN (O15091) Hypothetical protein KIAA0391 32 3.9
ODO2_BUCBP (Q89AJ6) Dihydrolipoyllysine-residue succinyltransfer... 32 3.9
YEFI_ECOLI (P37751) Hypothetical protein yefI 32 5.1
NU5M_CAEEL (P24896) NADH-ubiquinone oxidoreductase chain 5 (EC 1... 32 5.1
YE28_METJA (Q58823) Hypothetical protein MJ1428 31 6.7
RIR2_ASFM2 (P26713) Ribonucleoside-diphosphate reductase small c... 31 6.7
KDTA_RICPR (Q9ZE58) 3-deoxy-D-manno-octulosonic-acid transferase... 31 6.7
AROE_BUCAP (P46240) Shikimate dehydrogenase (EC 1.1.1.25) 31 6.7
>YLE1_SCHPO (Q10451) Hypothetical protein C1093.01 in chromosome I
Length = 1261
Score = 66.6 bits (161), Expect = 1e-10
Identities = 65/353 (18%), Positives = 144/353 (40%), Gaps = 50/353 (14%)
Query: 161 FHDKVLALGFQLNQVSYRTLINGLCKVGQTK---AALEMLRRIDGKLVRLDVVMYNTIID 217
F D + LG+ ++ LIN + G T AL + V+ V +YN ++
Sbjct: 875 FCDNMKMLGYIPRASTFAHLINNSTRRGDTDDATTALNIFEETKRHNVKPSVFLYNAVLS 934
Query: 218 GVCKDKLVNDAFDFYSEMVAKRICPTVVTYNTLICGLCIMGQLKDAIGLLHKMILENINP 277
+ + + + + + EM + PT VTY T+I
Sbjct: 935 KLGRARRTTECWKLFQEMKESGLLPTSVTYGTVI-------------------------- 968
Query: 278 TVYTFSILVDAFCKEGKVKEAKNVFVVMMKK-DVKPNIVTYNSLMNGYCLVNEVNKAESI 336
+A C+ G A+ +F M + + +P + YN+++ + + N+ +++
Sbjct: 969 ---------NAACRIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQ-FEVQTMFNREKAL 1018
Query: 337 F--NTMAQIGVAPDVHSYSIMISGFCKIKMVD-----EAMKLFEEMHCKQIFPDVVTYNS 389
F N + + P H+Y +++ + +K V+ ++L E + Y
Sbjct: 1019 FYYNRLCATDIEPSSHTYKLLMDAYGTLKPVNVGSVKAVLELMERTDVPILSMHYAAYIH 1078
Query: 390 LIDGLCKSGRISYALKLIG-EMHDRGQ-PPNIITYNSLLDALCKNHHVDKAIELLTKLKD 447
++ + + + + + HD G+ + + S +++L N + + I++++ +K
Sbjct: 1079 ILGNVVSDVQAATSCYMNALAKHDAGEIQLDANLFQSQIESLIANDRIVEGIQIVSDMKR 1138
Query: 448 HNIQPSVCTYNILINGLCKSGRLKDAQKVFEDVLVNGYN-IDVYTYNTMIKGF 499
+N+ + N LI G K G + A+ F+ + G + + TY M++ +
Sbjct: 1139 YNVSLNAYIVNALIKGFTKVGMISKARYYFDLLECEGMSGKEPSTYENMVRAY 1191
Score = 64.3 bits (155), Expect = 7e-10
Identities = 51/185 (27%), Positives = 89/185 (47%), Gaps = 9/185 (4%)
Query: 284 ILVDAFCKEGKVKEAKNVFVVMMKK-DVKPNIVTYNSLMNGYCL---VNEVNKAESIFNT 339
IL +F ++ K + N+F MK P T+ L+N ++ A +IF
Sbjct: 860 ILSSSFARQFK---SSNLFCDNMKMLGYIPRASTFAHLINNSTRRGDTDDATTALNIFEE 916
Query: 340 MAQIGVAPDVHSYSIMISGFCKIKMVDEAMKLFEEMHCKQIFPDVVTYNSLIDGLCKSGR 399
+ V P V Y+ ++S + + E KLF+EM + P VTY ++I+ C+ G
Sbjct: 917 TKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVINAACRIGD 976
Query: 400 ISYALKLIGEMHDR-GQPPNIITYNSLLDALCKN-HHVDKAIELLTKLKDHNIQPSVCTY 457
S A KL EM ++ P + YN+++ + + +KA+ +L +I+PS TY
Sbjct: 977 ESLAEKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNREKALFYYNRLCATDIEPSSHTY 1036
Query: 458 NILIN 462
+L++
Sbjct: 1037 KLLMD 1041
Score = 61.6 bits (148), Expect = 5e-09
Identities = 60/277 (21%), Positives = 114/277 (40%), Gaps = 14/277 (5%)
Query: 242 PTVVTYNTLICGLCIMGQLKDA---IGLLHKMILENINPTVYTFSILVDAFCKEGKVKEA 298
P T+ LI G DA + + + N+ P+V+ ++ ++ + + E
Sbjct: 886 PRASTFAHLINNSTRRGDTDDATTALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTEC 945
Query: 299 KNVFVVMMKKDVKPNIVTYNSLMNGYCLVNEVNKAESIFNTMA-QIGVAPDVHSYSIMIS 357
+F M + + P VTY +++N C + + + AE +F M Q P V Y+ MI
Sbjct: 946 WKLFQEMKESGLLPTSVTYGTVINAACRIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQ 1005
Query: 358 GFCKIKM-VDEAMKLFEEMHCKQIFPDVVTYNSLID--GLCKSGRISYALKLIGEMHDRG 414
+ ++A+ + + I P TY L+D G K + ++K + E+ +R
Sbjct: 1006 FEVQTMFNREKALFYYNRLCATDIEPSSHTYKLLMDAYGTLKPVNVG-SVKAVLELMERT 1064
Query: 415 QPPNIITYNSLLDALCKNHHVDKA------IELLTKLKDHNIQPSVCTYNILINGLCKSG 468
P + + + + N D + L K IQ + I L +
Sbjct: 1065 DVPILSMHYAAYIHILGNVVSDVQAATSCYMNALAKHDAGEIQLDANLFQSQIESLIAND 1124
Query: 469 RLKDAQKVFEDVLVNGYNIDVYTYNTMIKGFCKKGFV 505
R+ + ++ D+ +++ Y N +IKGF K G +
Sbjct: 1125 RIVEGIQIVSDMKRYNVSLNAYIVNALIKGFTKVGMI 1161
Score = 61.2 bits (147), Expect = 6e-09
Identities = 81/366 (22%), Positives = 159/366 (43%), Gaps = 23/366 (6%)
Query: 98 NGIASNLVTLSILINCFSQLGHNSLSFSVFSNILKKGYEPDAITLTTLIKGLCLKGDIH- 156
N + ++ IL + F++ +S F N+ GY P A T LI +GD
Sbjct: 849 NWFMALILDAMILSSSFARQFKSSNLFC--DNMKMLGYIPRASTFAHLINNSTRRGDTDD 906
Query: 157 --KALHFHDKVLALGFQLNQVSYRTLINGLCKVGQTKAALEMLRRIDGKLVRLDVVMYNT 214
AL+ ++ + + Y +++ L + +T ++ + + + V Y T
Sbjct: 907 ATTALNIFEETKRHNVKPSVFLYNAVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGT 966
Query: 215 IIDGVCKDKLVNDAFDFYSEMVAK-RICPTVVTYNTLI-CGLCIMGQLKDAIGLLHKMIL 272
+I+ C+ + A ++EM + P V YNT+I + M + A+ +++
Sbjct: 967 VINAACRIGDESLAEKLFAEMENQPNYQPRVAPYNTMIQFEVQTMFNREKALFYYNRLCA 1026
Query: 273 ENINPTVYTFSILVDAF--CKEGKVKEAKNVFVVMMKKDV---KPNIVTYNSLMNGYCLV 327
+I P+ +T+ +L+DA+ K V K V +M + DV + Y ++ +V
Sbjct: 1027 TDIEPSSHTYKLLMDAYGTLKPVNVGSVKAVLELMERTDVPILSMHYAAYIHILGN--VV 1084
Query: 328 NEVNKAESIF-NTMAQIG---VAPDVHSYSIMISGFCKIKMVDEAMKLFEEMHCKQIFPD 383
++V A S + N +A+ + D + + I + E +++ +M + +
Sbjct: 1085 SDVQAATSCYMNALAKHDAGEIQLDANLFQSQIESLIANDRIVEGIQIVSDMKRYNVSLN 1144
Query: 384 VVTYNSLIDGLCKSGRIS---YALKLIGEMHDRGQPPNIITYNSLLDALCKNHHVDKAIE 440
N+LI G K G IS Y L+ G+ P+ TY +++ A + KA+E
Sbjct: 1145 AYIVNALIKGFTKVGMISKARYYFDLLECEGMSGKEPS--TYENMVRAYLSVNDGRKAME 1202
Query: 441 LLTKLK 446
++ +LK
Sbjct: 1203 IVEQLK 1208
Score = 53.1 bits (126), Expect = 2e-06
Identities = 52/225 (23%), Positives = 92/225 (40%), Gaps = 31/225 (13%)
Query: 295 VKEAKNVFVVMMKKDVKPNIVTYNSLMNGYCLVNEVNKAESIFNTMAQIGVAPDVHSYSI 354
+ ++ N V M P V N L + + E NK + P+V Y
Sbjct: 766 IDQSINSLVSKMLTSASPEQVDVNILFFQFGKLIETNKF-----------LHPEV--YPT 812
Query: 355 MISGFCKIKMVDEAMKLFEEMHCKQIFPDVVTYNS---------LIDGLCKSGRISYALK 405
+IS K K D ++FE H K ++ + T + ++D + S + K
Sbjct: 813 LISVLSKNKRFDAVQRVFE--HSKHLYRKISTKSLEKANWFMALILDAMILSSSFARQFK 870
Query: 406 ----LIGEMHDRGQPPNIITYNSLLDALCKNHHVDKA---IELLTKLKDHNIQPSVCTYN 458
M G P T+ L++ + D A + + + K HN++PSV YN
Sbjct: 871 SSNLFCDNMKMLGYIPRASTFAHLINNSTRRGDTDDATTALNIFEETKRHNVKPSVFLYN 930
Query: 459 ILINGLCKSGRLKDAQKVFEDVLVNGYNIDVYTYNTMIKGFCKKG 503
+++ L ++ R + K+F+++ +G TY T+I C+ G
Sbjct: 931 AVLSKLGRARRTTECWKLFQEMKESGLLPTSVTYGTVINAACRIG 975
>LPRC_HUMAN (P42704) 130 kDa leucine-rich protein (LRP 130) (GP130)
(Leucine-rich PPR-motif containing protein)
Length = 1273
Score = 65.1 bits (157), Expect = 4e-10
Identities = 56/242 (23%), Positives = 105/242 (43%), Gaps = 5/242 (2%)
Query: 208 DVVMYNTIIDGVCKDKLVNDAFDFYSEMVAKRICPTVVTYNTLICGLCIMGQLKDAIGLL 267
DV YN ++ +++ DF ++M I P VTY LI C +G ++ A +L
Sbjct: 40 DVSHYNALLKVYLQNEYKFSPTDFLAKMEEANIQPNRVTYQRLIASYCNVGDIEGASKIL 99
Query: 268 HKMILENINPTVYTFSILVDAFCKEGKVKEAKNVFVVMMKKDVKPNIVTYNSLMNGYCLV 327
M +++ T FS LV + G ++ A+N+ VM ++P TY +L+N Y
Sbjct: 100 GFMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAGIEPGPDTYLALLNAYAEK 159
Query: 328 NEVNKAESIFNTMAQIGVAPDVHSYSIMISGFCKIKMVDEAMKLFEEMHC-KQIFPDVVT 386
+++ + + + + +I F K + + K +++ C ++ PD +
Sbjct: 160 GDIDHVKQTLEKVEKFELHLMDRDLLQIIFSFSKAGYLSMSQKFWKKFTCERRYIPDAM- 218
Query: 387 YNSLIDGLCKSGRISYALK-LIGEMHDRGQPPNIITYNSLLDALCKNHHVDKAIELLTKL 445
+LI L AL+ L+ + P++ L + N V+K + KL
Sbjct: 219 --NLILLLVTEKLEDVALQILLACPVSKEDGPSVFGSFFLQHCVTMNTPVEKLTDYCKKL 276
Query: 446 KD 447
K+
Sbjct: 277 KE 278
Score = 63.2 bits (152), Expect = 2e-09
Identities = 46/194 (23%), Positives = 85/194 (43%), Gaps = 2/194 (1%)
Query: 319 SLMNGYCLVNEVNKAESIFNTMAQIGVAPDVHSYSIMISGFCKIKMVDEAMKLFEEMHCK 378
SL+ L A I++T+ ++G DV Y+ ++ + + + +M
Sbjct: 11 SLLPELKLEERTEFAHRIWDTLQKLGAVYDVSHYNALLKVYLQNEYKFSPTDFLAKMEEA 70
Query: 379 QIFPDVVTYNSLIDGLCKSGRISYALKLIGEMHDRGQPPNIITYNSLLDALCKNHHVDKA 438
I P+ VTY LI C G I A K++G M + P +++L+ + ++ A
Sbjct: 71 NIQPNRVTYQRLIASYCNVGDIEGASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENA 130
Query: 439 IELLTKLKDHNIQPSVCTYNILINGLCKSGRLKDAQKVFEDVLVNGYNIDVYTYNTMIKG 498
+LT ++D I+P TY L+N + G + ++ E V ++ +I
Sbjct: 131 ENILTVMRDAGIEPGPDTYLALLNAYAEKGDIDHVKQTLEKVEKFELHLMDRDLLQIIFS 190
Query: 499 FCKKGF--VIPQFW 510
F K G+ + +FW
Sbjct: 191 FSKAGYLSMSQKFW 204
Score = 37.0 bits (84), Expect = 0.12
Identities = 61/335 (18%), Positives = 131/335 (38%), Gaps = 28/335 (8%)
Query: 76 LGSLVKANHYSIVVSLHRQMEFN-------------GIASNLVTLSILINCFSQLGHNSL 122
LG++ +HY+ ++ ++ Q E+ I N VT LI + +G
Sbjct: 35 LGAVYDVSHYNALLKVYLQNEYKFSPTDFLAKMEEANIQPNRVTYQRLIASYCNVGDIEG 94
Query: 123 SFSVFSNILKKGYEPDAITLTTLIKGLCLKGDIHKALHFHDKVLALGFQLNQVSYRTLIN 182
+ + + K + L+ G GD+ A + + G + +Y L+N
Sbjct: 95 ASKILGFMKTKDLPVTEAVFSALVTGHARAGDMENAENILTVMRDAGIEPGPDTYLALLN 154
Query: 183 GLCKVG---QTKAALEMLRRIDGKLVRLDVVMYNTIIDGVCKDKLVNDAFDFYSEMVA-K 238
+ G K LE + + + L+ D++ II K ++ + F+ + +
Sbjct: 155 AYAEKGDIDHVKQTLEKVEKFELHLMDRDLLQ---IIFSFSKAGYLSMSQKFWKKFTCER 211
Query: 239 RICPTVVTYNTLICGLCIMGQLKDAI--GLLHKMILENINPTVYTFSILVDAFCKEGKVK 296
R P + N ++ L + +L+D LL + + P+V+ L V+
Sbjct: 212 RYIPDAM--NLIL--LLVTEKLEDVALQILLACPVSKEDGPSVFGSFFLQHCVTMNTPVE 267
Query: 297 EAKNVFVVMMKKDVKPNIVTYNSLMNGYCLVNEVNKAESIFNTMAQIGVAPDVHSYSIMI 356
+ + + K+V+ + ++ L N+ + A+++ + + G H + ++
Sbjct: 268 KLTDYCKKL--KEVQMHSFPLQFTLHCALLANKTDLAKALMKAVKEEGFPIRPHYFWPLL 325
Query: 357 SGFCKIKMVDEAMKLFEEMHCKQIFPDVVTYNSLI 391
G K K V +++ + M + PD TY +
Sbjct: 326 VGRRKEKNVQGIIEILKGMQELGVHPDQETYTDYV 360
Score = 32.7 bits (73), Expect = 2.3
Identities = 48/224 (21%), Positives = 92/224 (40%), Gaps = 18/224 (8%)
Query: 125 SVFSNILKKGYEPDAITLTTLIKGLCLKGDIHKALHFHDKVLALGFQLNQVSYRTLINGL 184
+ +L + P + +T +I+ L +KGD+ + L + +S IN +
Sbjct: 1004 TTLKTVLDQQQTPSRLAVTRVIQALAMKGDVENIEVVQKMLNGLEDSIG-LSKMVFINNI 1062
Query: 185 C----KVGQTKAALEMLRRI---DGKLVRLDVVMYNTIIDGVCKDKLVNDAFDFYSEMVA 237
K AA+E + + + K++ + V +++L A + S M A
Sbjct: 1063 ALAQIKNNNIDAAIENIENMLTSENKVIEPQYFGLAYLFRKVIEEQL-EPAVEKISIM-A 1120
Query: 238 KRICPTVVTYNTL---ICGLCIMGQLKDAIGLLHKM-ILENINPTVYTFSILVDAFCKEG 293
+R+ Y + L G++ DA LL + + P + F L+ K+G
Sbjct: 1121 ERLANQFAIYKPVTDFFLQLVDAGKVDDARALLQRCGAIAEQTPILLLF--LLRNSRKQG 1178
Query: 294 KVKEAKNVFVVMMKKDVKPNIVTYNSLMNGYCLVNEVNKAESIF 337
K K+V ++ + + K YNSLM Y +V A++++
Sbjct: 1179 KASTVKSVLELIPELNEKEE--AYNSLMKSYVSEKDVTSAKALY 1220
Score = 32.0 bits (71), Expect = 3.9
Identities = 24/105 (22%), Positives = 49/105 (45%), Gaps = 5/105 (4%)
Query: 247 YNTLICGLCIMGQLKDAIGLLHKMILENINPTVYTFSIL--VDAFCKEGKVKEAKNVFVV 304
Y LI C +++DA+ L + + + + T + L V K GK+++A +
Sbjct: 593 YAALINLCCRHDKVEDALNLKEEFDRLDSSAVLDTGNYLGLVRVLAKHGKLQDAIKILKE 652
Query: 305 MMKKDV---KPNIVTYNSLMNGYCLVNEVNKAESIFNTMAQIGVA 346
M +KDV +++ ++NG L E+ + + + +G+A
Sbjct: 653 MKEKDVLIKDTTALSFFHMLNGAALRGEIETVKQLHEAIVTLGLA 697
Score = 31.2 bits (69), Expect = 6.7
Identities = 29/163 (17%), Positives = 68/163 (40%), Gaps = 19/163 (11%)
Query: 290 CKEGKVKEAKNVFVVMMKKDVKPNIVTYNSLMNGYCLVNEVNKAESIFNTMAQIGVAPDV 349
C+ + K A ++F+ ++++ N TY++L+ ++ +E F ++ +
Sbjct: 922 CRLNQKKGAYDIFLNAKEQNIVFNAETYSNLI-------KLLMSEDYFTQAMEVKAFAET 974
Query: 350 HSYSIMISGFCKIKMV---------DEAMKLFEEMHCKQIFPDVVTYNSLIDGLCKSG-- 398
H ++ +++ EA+ + + +Q P + +I L G
Sbjct: 975 HIKGFTLNDAANSRLIITQVRRDYLKEAVTTLKTVLDQQQTPSRLAVTRVIQALAMKGDV 1034
Query: 399 -RISYALKLIGEMHDRGQPPNIITYNSLLDALCKNHHVDKAIE 440
I K++ + D ++ N++ A KN+++D AIE
Sbjct: 1035 ENIEVVQKMLNGLEDSIGLSKMVFINNIALAQIKNNNIDAAIE 1077
>PT09_YEAST (P32522) PET309 protein, mitochondrial precursor
Length = 965
Score = 44.7 bits (104), Expect = 6e-04
Identities = 54/273 (19%), Positives = 115/273 (41%), Gaps = 14/273 (5%)
Query: 242 PTVVTYNTLICGLCIMGQLKDAIGLLHKMILENINPTVYTFSILVDAFCKEGKVKEAKNV 301
P++ Y L+ + +G+L L +++ + PT L+ A K G +
Sbjct: 344 PSIQHYGILMYTMARIGELDSVNKLYTQLLRRGMIPTYAVLQSLLYAHYKVGDFAACFSH 403
Query: 302 FVVMMKKDVKPNIVTYNSLMNGYCLVNEVNKAESIFNTMAQIGVAPDVH----SYSIMIS 357
F + K D+ P+ T+ ++ Y +N+++ A F + ++ P V ++++I
Sbjct: 404 FELFKKYDITPSTATHTIMLKVYRGLNDLDGA---FRILKRLSEDPSVEITEGHFALLIQ 460
Query: 358 GFCKIKMVDEAMKLFEEM-HCKQIFPDVVTYNSLIDGLCKSGRISYALKLIGEMHDRG-- 414
CK A +LF M I + ++L+D +S R + A+ L E H +
Sbjct: 461 MCCKTTNHLIAQELFNLMTEHYNIQHTGKSISALMDVYIESNRPTEAIALF-EKHSKNLS 519
Query: 415 -QPPNIITYNSLLDALCKNHHVDKAIELLTKLKDHNIQPSVCTYNILINGLCKSGR-LKD 472
+ I YN + A + +K EL K+ + + Y ++I L +
Sbjct: 520 WRDGLISVYNKAIKAYIGLRNANKCEELFDKITTSKLAVNSEFYKMMIKFLVTLNEDCET 579
Query: 473 AQKVFEDVLVNG-YNIDVYTYNTMIKGFCKKGF 504
A + + ++ + +D + +++ + K+G+
Sbjct: 580 ALSIIDQLIKHSVIKVDATHFEIIMEAYDKEGY 612
Score = 43.9 bits (102), Expect = 0.001
Identities = 35/172 (20%), Positives = 76/172 (43%), Gaps = 2/172 (1%)
Query: 317 YNSLMNGYCLVNEVNKAESIFNTMAQIGVAPDVHSYSIMISGFCKIKMVDEAMKLFEEMH 376
++ L+ + ++ + + FN + IG P + Y I++ +I +D KL+ ++
Sbjct: 314 FDYLLVAHSRLHNWDALQQQFNALFGIGKLPSIQHYGILMYTMARIGELDSVNKLYTQLL 373
Query: 377 CKQIFPDVVTYNSLIDGLCKSGRISYALKLIGEMHDRGQPPNIITYNSLLDALCKNHHVD 436
+ + P SL+ K G + P+ T+ +L + +D
Sbjct: 374 RRGMIPTYAVLQSLLYAHYKVGDFAACFSHFELFKKYDITPSTATHTIMLKVYRGLNDLD 433
Query: 437 KAIELLTKL-KDHNIQPSVCTYNILINGLCKSGRLKDAQKVFEDVLVNGYNI 487
A +L +L +D +++ + + +LI CK+ AQ++F +++ YNI
Sbjct: 434 GAFRILKRLSEDPSVEITEGHFALLIQMCCKTTNHLIAQELF-NLMTEHYNI 484
Score = 32.3 bits (72), Expect = 3.0
Identities = 27/142 (19%), Positives = 52/142 (36%), Gaps = 2/142 (1%)
Query: 320 LMNGYCLVNEVNKAESIFNTMAQIGVAPDVHSYSIMISGFCKIKMVDEAMKLFEEMHCKQ 379
++ YC + + N S + D + + ++ ++ D + F +
Sbjct: 284 ILKTYCHMQKFNLVSSTYWKYPD--AQHDQNQFDYLLVAHSRLHNWDALQQQFNALFGIG 341
Query: 380 IFPDVVTYNSLIDGLCKSGRISYALKLIGEMHDRGQPPNIITYNSLLDALCKNHHVDKAI 439
P + Y L+ + + G + KL ++ RG P SLL A K
Sbjct: 342 KLPSIQHYGILMYTMARIGELDSVNKLYTQLLRRGMIPTYAVLQSLLYAHYKVGDFAACF 401
Query: 440 ELLTKLKDHNIQPSVCTYNILI 461
K ++I PS T+ I++
Sbjct: 402 SHFELFKKYDITPSTATHTIML 423
Score = 31.6 bits (70), Expect = 5.1
Identities = 23/99 (23%), Positives = 43/99 (43%), Gaps = 19/99 (19%)
Query: 356 ISGFCKIKMVDEAMKLFEEMHCKQIFPDVVTYNSLIDGLCKSGR-ISYALKLIGEMHDRG 414
+S + +DE L++++ K D +++NS ++ L K R +SY +K++
Sbjct: 782 VSQLIAMNKIDELPLLWKKLREKGFILDNISWNSAVEALFKDPRTLSYGMKIVD------ 835
Query: 415 QPPNIITYNSLLDALCKNHHVDKAIELLTKLKDHNIQPS 453
D L +++ LLTKL + Q S
Sbjct: 836 ------------DTLIHGYNLIHKFRLLTKLSEDKTQSS 862
>YG3M_YEAST (P48237) Hypothetical 101.4 kDa protein in RPL24B-RSR1
intergenic region
Length = 864
Score = 42.4 bits (98), Expect = 0.003
Identities = 19/51 (37%), Positives = 32/51 (62%)
Query: 417 PNIITYNSLLDALCKNHHVDKAIELLTKLKDHNIQPSVCTYNILINGLCKS 467
P+I TYN++L K + KA++L +++DHNI+P+ TY ++ L S
Sbjct: 355 PDICTYNTMLRICEKERNFPKALDLFQEIQDHNIKPTTNTYIMMARVLASS 405
Score = 37.7 bits (86), Expect = 0.071
Identities = 22/81 (27%), Positives = 38/81 (46%), Gaps = 2/81 (2%)
Query: 344 GVAPDVHSYSIMISGFCKIKMVDEAMKLFEEMH--CKQIFPDVVTYNSLIDGLCKSGRIS 401
G+ P+ + + +I + + +M +A F+ M + FPD+ TYN+++ K
Sbjct: 315 GITPNKQNLTTVIQFYSRKEMTKQAWNTFDTMKFLSTKHFPDICTYNTMLRICEKERNFP 374
Query: 402 YALKLIGEMHDRGQPPNIITY 422
AL L E+ D P TY
Sbjct: 375 KALDLFQEIQDHNIKPTTNTY 395
>YEQ7_YEAST (P40050) Hypothetical 79.5 kDa protein in PTP3-SER3
intergenic region
Length = 688
Score = 38.5 bits (88), Expect = 0.042
Identities = 49/197 (24%), Positives = 77/197 (38%), Gaps = 36/197 (18%)
Query: 286 VDAFCKEGKVKEAKNVFVVMMKKDVKPNIVTYNSLMNGYC-LVNEVNKAESIFNTMAQIG 344
V F K G++++A VF+ + K K +V N +M Y +V A IFN + G
Sbjct: 174 VQKFLKRGQLEKA--VFLARLAK--KKGVVGMNLIMKYYIEVVQSQQSAVDIFNWRKKWG 229
Query: 345 VAPDVHSYSIMISGFCKIKMVDEAMKLFEEMHCKQIFPDVVTYNSLIDGLCKSGRISYAL 404
V D HS +I+ +G K + + K + E+ K ID LC ++
Sbjct: 230 VPIDQHSITILFNGLSKQENL--VSKKYGELVLK-----------TIDSLCDKNELTE-- 274
Query: 405 KLIGEMHDRGQPPNIITYNSLLDALCKNHHVDKAIELLTKLKDHNIQPSVCTYNILINGL 464
I YN+ L AL +LL K K ++ TY ++I
Sbjct: 275 ---------------IEYNTALAALINCTDETLVFKLLNK-KCPGLKKDSITYTLMIRSC 318
Query: 465 CKSGRLKDAQKVFEDVL 481
+ K V D++
Sbjct: 319 TRIADEKRFMVVLNDLM 335
>NMT_MSEPV (Q9YW20) Putative glycylpeptide
N-tetradecanoyltransferase (EC 2.3.1.97) (Peptide
N-myristoyltransferase) (Myristoyl-CoA:protein
N-myristoyltransferase) (NMT)
Length = 298
Score = 37.7 bits (86), Expect = 0.071
Identities = 48/242 (19%), Positives = 103/242 (41%), Gaps = 37/242 (15%)
Query: 111 INCFSQLGHNSLSFSVFSNILKKGYEPDAI--TLTTLIKGLCLKGDIHKALHFHDKVLAL 168
I+ FS L N S F+ +L Y+ D I T+ +++ + +K +IHK +H +
Sbjct: 67 IDTFSWLVLNPFSDPSFNIVL---YDNDKIVATIVGILRSIKIKNEIHKIIHTTFLTVDE 123
Query: 169 GFQLNQVSYRTLINGLCKVGQTKAAL-------EMLRRI------DGKLVRLDVVMY--- 212
++ + + +I+ L + K L + +++I D +++ D Y
Sbjct: 124 NYRKQGIHFY-IIDKLMENAFNKGVLLGIFSTMKKIKKIKCVNVQDTYIIKSDSKKYKEN 182
Query: 213 NTIIDGVCKDKLVNDAFDFYSEMVA-----KRICPTVVTYNTLICGLCIMGQLKDAIGLL 267
N + D ++ +D + Y++M K+ C + YN L C L I + + + +L
Sbjct: 183 NNVFDYKKLNQKNDDLYFIYNDMEIEYWFNKKYCHIISIYNNLFCFLKIKYKNNENLNIL 242
Query: 268 HKMILENINPTVYTF---SILVDAFCKEGKVKEAKNVFVVMMKKDVKPNIVTYNSLMNGY 324
+ + N N Y+ SI+ + + ++K ++ + + +N+L
Sbjct: 243 IEQYIHNKNINKYSIPNNSIMFSQYIQLPQIKLQNQIYTYIYN-------LNFNNLKCNI 295
Query: 325 CL 326
C+
Sbjct: 296 CM 297
>GLNA_BUTFI (Q05650) Glutamine synthetase (EC 6.3.1.2)
(Glutamate--ammonia ligase) (GS)
Length = 700
Score = 35.0 bits (79), Expect = 0.46
Identities = 22/86 (25%), Positives = 46/86 (52%), Gaps = 5/86 (5%)
Query: 261 KDAIGLLHKMILENINPTVYTFSILVDAFCKEGKVKEAKNVFVVMMKKDVKPNIVTYNSL 320
K+ I ++++LEN Y+ SI +++ + +++ +V +KD+ IV SL
Sbjct: 556 KEEIYSRYEILLEN-----YSKSIHIESLTMQEMIRKDLTEGLVAYEKDLSKEIVQKKSL 610
Query: 321 MNGYCLVNEVNKAESIFNTMAQIGVA 346
++G C E+ +S+ + A++G A
Sbjct: 611 LDGDCCALELGVLKSLDKSSAEMGKA 636
>UAB2_PIG (P46202) Uteroferrin-associated basic protein 2 precursor
(UABP-2)
Length = 420
Score = 33.5 bits (75), Expect = 1.3
Identities = 48/198 (24%), Positives = 77/198 (38%), Gaps = 27/198 (13%)
Query: 267 LHKMILENINPTVYTFSILVDAFCKEGKVKEAKNVFVVMMKKDVKPNIVTYNSLMNGYCL 326
+++M L+ I+ VY I + F + K K+A N FV N++T+ CL
Sbjct: 150 MNQMFLKEID-RVYKVDIQMIDFKDKEKTKKAINQFVADKIDKKAKNLITHLDPQTLLCL 208
Query: 327 VNEV--------------NKAESIFNTMAQIGVAPDVHSYSIMISGFCKIKMVDEAMKLF 372
VN V K E F I + MI +E +
Sbjct: 209 VNYVFFKGILERAFQTNLTKKEDFFVNEKTIVQVDMMRKTERMI-----YSRSEELLATM 263
Query: 373 EEMHCKQIFPDVVTYNSLIDGLCKSGRISYALKLIGEMHDRGQPPNIITYNSLLDALCKN 432
+M CK+ S+I L +G+ +ALK + R Q N + +L A ++
Sbjct: 264 VKMPCKE-------NASIILVLPDTGKFDFALKEMAAKRARLQKTNELQIGALSCAQDQD 316
Query: 433 HHVDKAIELLTKLKDHNI 450
H D+ LL K+ ++I
Sbjct: 317 HLQDRFKHLLPKIGINDI 334
>POLB_MAIZE (P15718) Retrovirus-related Pol polyprotein from
transposon element BS1 (ORF 1) [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 740
Score = 33.5 bits (75), Expect = 1.3
Identities = 20/67 (29%), Positives = 33/67 (48%), Gaps = 9/67 (13%)
Query: 365 VDEAMKLFEEMHCKQ---------IFPDVVTYNSLIDGLCKSGRISYALKLIGEMHDRGQ 415
V++A K+F++M K + V +N L+ L GR S A ++ EM + G
Sbjct: 442 VEDAAKVFDKMLTKGEEAPSVQRGVVMTAVAHNLLVQALFMDGRASDAYVVLEEMQNNGP 501
Query: 416 PPNIITY 422
P++ TY
Sbjct: 502 FPDVFTY 508
Score = 33.5 bits (75), Expect = 1.3
Identities = 22/73 (30%), Positives = 35/73 (47%), Gaps = 9/73 (12%)
Query: 433 HHVDKAIELLTKLKDHNIQ-PSV--------CTYNILINGLCKSGRLKDAQKVFEDVLVN 483
+HV+ A ++ K+ + PSV +N+L+ L GR DA V E++ N
Sbjct: 440 YHVEDAAKVFDKMLTKGEEAPSVQRGVVMTAVAHNLLVQALFMDGRASDAYVVLEEMQNN 499
Query: 484 GYNIDVYTYNTMI 496
G DV+TY +
Sbjct: 500 GPFPDVFTYTLKV 512
>ATF2_YEAST (P53296) Alcohol O-acetyltransferase 2 (EC 2.3.1.84)
(AATase 2)
Length = 535
Score = 33.5 bits (75), Expect = 1.3
Identities = 19/47 (40%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Query: 15 ITNFPNFQFLNPIFLIHCFTSYSSFSNSTTLYSQLHNLVSSFNHLLH 61
+T+ P F FL HC SS S T YS N +S+N+LLH
Sbjct: 255 LTSLPKF-FLTTFIYEHCNFKTSSESTLTARYSPSSNANASYNYLLH 300
>XAB1_MOUSE (Q8VCE2) XPA-binding protein 1
Length = 372
Score = 33.1 bits (74), Expect = 1.8
Identities = 24/111 (21%), Positives = 57/111 (50%), Gaps = 7/111 (6%)
Query: 390 LIDGLCKSGRISYALKLIGEMHDRGQPPNIITYNSLLDALCKNHHVD--KAIELLTKLKD 447
L+ G+ SG+ ++ +L G +H++G PP +I + + + ++D ++ +K
Sbjct: 23 LVLGMAGSGKTTFVQRLTGHLHNKGCPPYVINLDPAVHEVPFPANIDIRDTVKYKEVMKQ 82
Query: 448 HNIQPS---VCTYNILINGLCKSGR-LKDAQKVFEDVLVN-GYNIDVYTYN 493
+ + P+ V + N+ + + ++ AQ F VL++ I+V+T++
Sbjct: 83 YGLGPNGGIVTSLNLFATRFDQVMKFIEKAQNTFRYVLIDTPGQIEVFTWS 133
>INB_HUMAN (P01574) Interferon beta precursor (IFN-beta) (Fibroblast
interferon)
Length = 187
Score = 32.3 bits (72), Expect = 3.0
Identities = 26/97 (26%), Positives = 45/97 (45%), Gaps = 4/97 (4%)
Query: 21 FQFLNPIFLIHCFTSYSSFSNSTTLYSQLHNLVSSFNHL---LHQKNPTPSIIQFGKILG 77
++ L IF I S S+ N T + + L N+ NHL L +K + GK++
Sbjct: 81 YEMLQNIFAIFRQDSSSTGWNETIVENLLANVYHQINHLKTVLEEKLEKEDFTR-GKLMS 139
Query: 78 SLVKANHYSIVVSLHRQMEFNGIASNLVTLSILINCF 114
SL +Y ++ + E++ A +V + IL N +
Sbjct: 140 SLHLKRYYGRILHYLKAKEYSHCAWTIVRVEILRNFY 176
>Y391_HUMAN (O15091) Hypothetical protein KIAA0391
Length = 567
Score = 32.0 bits (71), Expect = 3.9
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 1/70 (1%)
Query: 177 YRTLINGLCKVGQTKAALEMLRRIDGKLVRLDVVMYNTIIDGVCKDKLVNDAFDFYSEMV 236
Y LI GL + + AL +L I K++ YN I G + VN A++ Y E++
Sbjct: 213 YSLLIRGLIHSDRWREALLLLEDIK-KVITPSKKNYNDCIQGALLHQDVNTAWNLYQELL 271
Query: 237 AKRICPTVVT 246
I P + T
Sbjct: 272 GHDIVPMLET 281
>ODO2_BUCBP (Q89AJ6) Dihydrolipoyllysine-residue succinyltransferase
component of 2-oxoglutarate dehydrogenase complex (EC
2.3.1.61) (E2) (Dihydrolipoamide succinyltransferase
component of 2-oxoglutarate dehydrogenase complex)
Length = 410
Score = 32.0 bits (71), Expect = 3.9
Identities = 31/130 (23%), Positives = 57/130 (43%), Gaps = 5/130 (3%)
Query: 261 KDAIGLLHKMILENINPTVYTFSILVDAFCKEGKV-KEAKNVFVVMMKKDVKPNIVTYNS 319
KD + L K I N N + + + F K + K V + ++K + +++ +
Sbjct: 144 KDILNYL-KNIRSNTNKKINNYDLNAYNFNTTHKNHRSIKRVKMTRLRKKISERLLSTKN 202
Query: 320 LMNGYCLVNEVNKAESIFNTMAQIG-VAPDVHSYSIMISGFCKIKMVDEAMKLFEEMHCK 378
NEVN +SI N + G + H + + F +K V EA+K+F E++
Sbjct: 203 NTASLTTFNEVNM-QSILNLRRKYGELFKQKHGIKLGLMSFY-VKAVIEALKIFPEINAS 260
Query: 379 QIFPDVVTYN 388
+++ YN
Sbjct: 261 IDNDEIIYYN 270
>YEFI_ECOLI (P37751) Hypothetical protein yefI
Length = 372
Score = 31.6 bits (70), Expect = 5.1
Identities = 37/157 (23%), Positives = 64/157 (40%), Gaps = 25/157 (15%)
Query: 12 SVFITNFPNFQFLNPIFLIHCFTSYSSFSNSTTLYSQLHNLVSSFNHLLHQKNPTPSIIQ 71
S+ + + NF P ++ F + ++ + + + +H+ L + P I+
Sbjct: 4 SIVVVSAVNFTTGGPFTILKKFLAATNNKENVSFIALVHSAKE-----LKESYPWVKFIE 58
Query: 72 FGKILGSLVKANHYSIVVSLHRQMEFNG--------IASNLVTLSILINCFSQLGHNSLS 123
F ++ GS +K H+ VV E N I +N+VT + C HN
Sbjct: 59 FPEVKGSWLKRLHFEYVVCKKLSKELNATHWICLHDITANVVTKKRYVYC-----HNPAP 113
Query: 124 F---SVFSNILKKGYEPDAITLTTLIKGLCLKGDIHK 157
F +F IL EP + L ++ GL K +I K
Sbjct: 114 FYKGILFREIL---MEP-SFFLFKMLYGLIYKINIKK 146
>NU5M_CAEEL (P24896) NADH-ubiquinone oxidoreductase chain 5 (EC
1.6.5.3)
Length = 527
Score = 31.6 bits (70), Expect = 5.1
Identities = 46/170 (27%), Positives = 76/170 (44%), Gaps = 22/170 (12%)
Query: 34 TSYSSFSNSTTLYSQLHNLVSSFNHLLHQKNPTPSII------QFGKILGSLVKANHYSI 87
T SS +S+TL + L+ +FN+L+ QK+ ++ F L SLV+ + +
Sbjct: 195 TPVSSLVHSSTLVTAGLILLMNFNNLVMQKDFISFVLIIGLFTMFFSSLASLVEEDLKKV 254
Query: 88 V-VSLHRQMEFNGIA----------SNLVTLSILINC-FSQLG---HNSLSFSVFSNILK 132
V +S QM F+ + +LV+ ++ +C F Q+G H S N
Sbjct: 255 VALSTLSQMGFSMVTLGLGLSFISFIHLVSHALFKSCLFMQVGYIIHCSFGQQDGRNYSN 314
Query: 133 KGYEPDAITLTTLIKGLCLKGDIHKA-LHFHDKVLALGFQLNQVSYRTLI 181
G P+ I L L+ CL G I + D +L L F N + + +L+
Sbjct: 315 NGNLPNFIQLQMLVTLFCLCGLIFSSGAVSKDFILELFFSNNYMMFFSLM 364
>YE28_METJA (Q58823) Hypothetical protein MJ1428
Length = 567
Score = 31.2 bits (69), Expect = 6.7
Identities = 44/178 (24%), Positives = 80/178 (44%), Gaps = 16/178 (8%)
Query: 152 KGDIHKALHFHDKVLALGFQ------LNQVSYRTLINGLCKVGQTKAALEMLRRIDGKLV 205
KG++ K+ +F DKVL + ++ ++ +LI G K E ++ +D L+
Sbjct: 315 KGELEKSSNFFDKVLETYLEELSEEDISALNLYSLIGKAETTGIPKYYHEAMKYVD-NLI 373
Query: 206 RLDVVMYNTIIDGVCKDKLVNDAFDFYSEMVAKRICP-TVVTYNTLICGLCIMGQLKDAI 264
L+ + G KL N + S M A R+ P + T +L L G++ +AI
Sbjct: 374 NLENSSRWWYVKGYIYYKLGNYKDAYESFMNALRVNPKDISTLKSLAIVLEKSGKIDEAI 433
Query: 265 GLLHKMILENINPTVYTFSILVDAFCKEGKVKEAKNVFVVMMKKDV-----KPNIVTY 317
K IL+ +N +T I D ++ + ++ N+ + + D+ KPNI +
Sbjct: 434 TTYTK-ILKIVNSLQFTCEI--DNILEKLRSRKPTNLEIPSVLFDIPVMYHKPNITCF 488
>RIR2_ASFM2 (P26713) Ribonucleoside-diphosphate reductase small
chain (EC 1.17.4.1) (Ribonucleotide reductase)
Length = 327
Score = 31.2 bits (69), Expect = 6.7
Identities = 33/155 (21%), Positives = 66/155 (42%), Gaps = 15/155 (9%)
Query: 311 KPNIVTYNSLMNGYCLVNEVNKAESIFNTMAQIGVAPDVHSYSIMISGFCKIKMVDEAMK 370
KP Y ++ + + +E+ + N M +I V ++ Y++ + C + EA
Sbjct: 53 KPQREFYKQILAFFVVADEIVIENLLTNFMREIKVKEVLYFYTMQAAQEC---VHSEAYS 109
Query: 371 LFEEMHCKQIFPDVVTYNSLIDGLCKSGRISYALKLIGEMHDRGQPP---NIITYNSLLD 427
+ K + PD + G+ K I + + + D + ++ + ++
Sbjct: 110 I----QVKTLIPDEKEQQRIFSGIEKHPIIKKMAQWVRQWMDPARNSLGERLVGFAAVEG 165
Query: 428 ALCKNHHVDKAIELLTKLKDHNIQPSVCTYNILIN 462
L +NH V AI+ L K+ NI P + +YN I+
Sbjct: 166 ILFQNHFV--AIQFL---KEQNIMPGLVSYNEFIS 195
>KDTA_RICPR (Q9ZE58) 3-deoxy-D-manno-octulosonic-acid transferase
(EC 2.-.-.-) (KDO transferase)
Length = 461
Score = 31.2 bits (69), Expect = 6.7
Identities = 18/51 (35%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query: 99 GIASNLVTLSILINCFSQLGHNSLSFSVFSNILKKGYEPDAITLTTLIKGL 149
G+ ++ T+S + F Q GHN L + FSN + G PD T + KG+
Sbjct: 351 GLFFSVATISFIGGSFKQGGHNILEAAYFSNCIIFG--PDMSKNTDIAKGI 399
>AROE_BUCAP (P46240) Shikimate dehydrogenase (EC 1.1.1.25)
Length = 273
Score = 31.2 bits (69), Expect = 6.7
Identities = 31/109 (28%), Positives = 49/109 (44%), Gaps = 11/109 (10%)
Query: 231 FYSEMVAKRICPTVVTYNTL--ICGLCIMGQLKDAIGLLHKMILENINPTVYTFSILVDA 288
F+S+ + +R + NTL I CI+G D IGLL ++ +N FSIL+
Sbjct: 75 FFSDKLTER-AKIAQSVNTLKKISDKCILGDNTDGIGLLSDLV--RLNFIKKNFSILI-- 129
Query: 289 FCKEGKVKEAKNVFVVMMKKDVKPNIVTYNSLMNGYCLVNEVNKAESIF 337
G K V + ++ I+ +++N LV + NK IF
Sbjct: 130 ---LGAGGAVKGVLLPLLSLGCSVYILN-RTILNAKILVKQFNKYGKIF 174
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.140 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 58,115,255
Number of Sequences: 164201
Number of extensions: 2412736
Number of successful extensions: 7761
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 7694
Number of HSP's gapped (non-prelim): 60
length of query: 512
length of database: 59,974,054
effective HSP length: 115
effective length of query: 397
effective length of database: 41,090,939
effective search space: 16313102783
effective search space used: 16313102783
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC147712.9


