

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
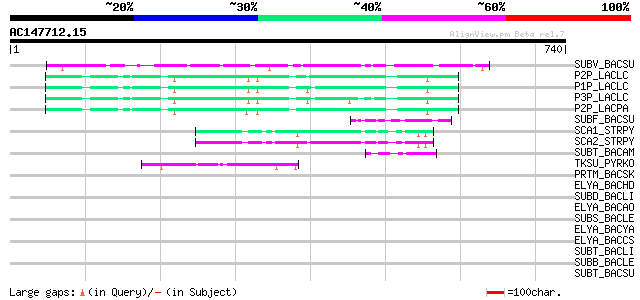
Query= AC147712.15 + phase: 0 /pseudo/partial
(740 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (... 122 3e-27
P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96) ... 84 1e-15
P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-) (W... 81 1e-14
P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)... 79 3e-14
P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96) ... 75 5e-13
SUBF_BACSU (P16397) Bacillopeptidase F precursor (EC 3.4.21.-) (... 51 1e-05
SCA1_STRPY (P15926) C5a peptidase precursor (EC 3.4.21.-) (SCP) 47 2e-04
SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP) 46 3e-04
SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62) (Su... 46 4e-04
TKSU_PYRKO (P58502) Tk-subtilisin precursor (EC 3.4.21.-) 45 5e-04
PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-) 44 0.001
ELYA_BACHD (P41363) Thermostable alkaline protease precursor (EC... 44 0.002
SUBD_BACLI (P00781) Subtilisin DY (EC 3.4.21.62) 43 0.003
ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-) 43 0.003
SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline... 42 0.004
ELYA_BACYA (P20724) Alkaline elastase YaB precursor (EC 3.4.21.-) 42 0.004
ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-) 42 0.004
SUBT_BACLI (P00780) Subtilisin Carlsberg precursor (EC 3.4.21.62) 42 0.006
SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline prote... 42 0.008
SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62) 41 0.013
>SUBV_BACSU (P29141) Minor extracellular protease vpr precursor (EC
3.4.21.-)
Length = 806
Score = 122 bits (307), Expect = 3e-27
Identities = 148/613 (24%), Positives = 255/613 (41%), Gaps = 98/613 (15%)
Query: 49 YNKHINGFAAVLEVEEAAKI-------AKHPNVVSVFEN-KGHELQTTRSWEFLGLENNY 100
Y + +GF+ L E K+ A +PNV +N K ++ + ++++
Sbjct: 106 YEQVFSGFSMKLPANEIPKLLAVKDVKAVYPNVTYKTDNMKDKDVTISEDAVSPQMDDSA 165
Query: 101 GVVPKDSIWEKGRYGEGTIIANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDNFHCNRK 160
+ + W+ G G+G +A ID+GV ++ + +++G +DN
Sbjct: 166 PYIGANDAWDLGYTGKGIKVAIIDTGVE-----YNHPDLKKNFGQYKGYDFVDN------ 214
Query: 161 LIGARFYSQGYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGL--ANGTAK 218
Y+ K TPT G G +V G ANGT K
Sbjct: 215 ---------DYDPK-----------------ETPTGDPRGEATDHGTHVAGTVAANGTIK 248
Query: 219 GGSPRSHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGI 278
G +P + + AY+V G T +++ E A+ DG D+++ SLG + +
Sbjct: 249 GVAPDATLLAYRVLGPGG---SGTTENVIAGVERAVQDGADVMNLSLGNSLNNPDWATST 305
Query: 279 SIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIM 338
++ A+ GV+ V GNSGP TV + +++ + L L + +
Sbjct: 306 ALD--WAMSEGVVAVTSNGNSGPNGWTVGSPGTSREAISVGA------TQLPLNEYAVTF 357
Query: 339 GTSLST---GLPNEKFYSLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKILFCLLREL 395
G+ S G E +++ + ++ A I +AK + + GK+ + R
Sbjct: 358 GSYSSAKVMGYNKEDDVKALNNKEVELVEAGIGEAK-----DFEGKDLTGKVAV-VKRGS 411
Query: 396 DGLVYAEEEAISGGSIGLVLGNDKQRGNDIMAYAHLLPTSHINYTDGEYVHSYIKATKTP 455
V + A G+IG+V+ N+ + +PT ++ DGE + S +KA +T
Sbjct: 412 IAFVDKADNAKKAGAIGMVVYNNLSGEIEANVPGMSVPTIKLSLEDGEKLVSALKAGETK 471
Query: 456 MAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLA 515
+ +G + +A SSRGP + ++KPDI+APGV+I+ + +
Sbjct: 472 TTFKLTVSKALGEQ----VADFSSRGP-VMDTWMIKPDISAPGVNIV---------STIP 517
Query: 516 SDNQWIPYNIGS--GTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPI 573
+ + PY GS GTS++ PH++ VA++K P WS K+AIM T +
Sbjct: 518 THDPDHPYGYGSKQGTSMASPHIAGAVAVIKQAKPKWSVEQIKAAIMNTAV-------TL 570
Query: 574 KDQSKEDATPFGYGAGHIQPELAMDPGLVYDLNIVDYLNFLCAHGYNQTQMKMFS----- 628
KD E GAG + A+ + Y FL +G N+T+ + F+
Sbjct: 571 KDSDGEVYPHNAQGAGSARIMNAIKADSLVSPGSYSYGTFLKENG-NETKNETFTIENQS 629
Query: 629 --RKPYICPKSYN 639
RK Y S+N
Sbjct: 630 SIRKSYTLEYSFN 642
>P2P_LACLC (P15293) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 84.0 bits (206), Expect = 1e-15
Identities = 133/598 (22%), Positives = 227/598 (37%), Gaps = 95/598 (15%)
Query: 48 SYNKHINGFAAVLEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDS 107
SY +NGF+ + V + K+ + V +V K + ++ ++ +
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMANVQ---------A 199
Query: 108 IWEKGRY-GEGTIIANIDSGVSPESKSF--SDDGMGPVPSRWRGICQLDNFHCNRKLIGA 164
+W +Y GEGT+++ IDSG+ P K SDD + ++ F K G
Sbjct: 200 VWSNYKYKGEGTVVSVIDSGIDPTHKDMRLSDDKDVKLTKS-----DVEKFTDTAKH-GR 253
Query: 165 RFYSQ-GYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAK---GG 220
F S+ Y + N ++ + HG + G N G + AK G
Sbjct: 254 YFNSKVPYGFNYADNNDTITDDTVDEQHGMHVAGIIGAN--------GTGDDPAKSVVGV 305
Query: 221 SPRSHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISI 280
+P + + A KV A ++ A ED+ G D+++ SLG S + ED
Sbjct: 306 APEAQLLAMKVFTNSDTSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGNQTLEDPELA 365
Query: 281 GAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSV-----------------AASTIDR 323
+A E+G V GNSG V + + AS +
Sbjct: 366 AVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGTPGTSRGATTVASAENT 425
Query: 324 NFVSY---------LQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICK 374
+ ++ LQLG + I + ++ TG ++K + +V + + D
Sbjct: 426 DVITQAVTITDGTGLQLGPETIQLSSNDFTGSFDQKKFYVVKDASGNLSKGKVADYTA-- 483
Query: 375 VGSLDPNKVKGKILFCLLRELDGLVYAEEE--AISGGSIGLVLGNDKQRGNDI--MAYAH 430
KGKI EL +A+++ A + G+ GL++ N+ + MA
Sbjct: 484 -------DAKGKIAIVKRGEL---TFADKQKYAQAAGAAGLIIVNNDGTATPVTSMALTT 533
Query: 431 LLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIIL 490
PT ++ G+ + ++ A + A T V + P+ +
Sbjct: 534 TFPTFGLSSVTGQKLVDWVAAHPDDSLGVKIALTLVPNQKYTEDKMSDFTSYGPVSNLSF 593
Query: 491 KPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNW 550
KPDITAPG +I S Y SGTS++ P ++ ALLK N
Sbjct: 594 KPDITAPGGNIW-------------STQNNNGYTNMSGTSMASPFIAGSQALLKQALNNK 640
Query: 551 SPAAF------KSAIMT--TTTIQGNNHRPIKDQSKED--ATPFGYGAGHIQPELAMD 598
+ + K +T T++ N +PI D + + +P GAG + + A+D
Sbjct: 641 NNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLVDVKAAID 698
>P1P_LACLC (P16271) PI-type proteinase precursor (EC 3.4.21.-)
(Wall-associated serine proteinase)
Length = 1902
Score = 80.9 bits (198), Expect = 1e-14
Identities = 133/599 (22%), Positives = 224/599 (37%), Gaps = 97/599 (16%)
Query: 48 SYNKHINGFAAVLEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDS 107
SY +NGF+ + V + K+ + V +V K + ++ ++ +
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMANVQ---------A 199
Query: 108 IWEKGRY-GEGTIIANIDSGVSPESKSF--SDDGMGPVPSRWRGICQLDNFHCNRKLIGA 164
+W +Y GEGT+++ IDSG+ P K SDD + ++ F K G
Sbjct: 200 VWSNYKYKGEGTVVSVIDSGIDPTHKDMRLSDDKDVKLTKS-----DVEKFTDTAKH-GR 253
Query: 165 RFYSQ-GYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAK---GG 220
F S+ Y + N ++ + HG + G N G + AK G
Sbjct: 254 YFNSKVPYGFNYADNNDTITDDTVDEQHGMHVAGIIGAN--------GTGDDPAKSVVGV 305
Query: 221 SPRSHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISI 280
+P + + A KV + ++ A ED+ G D+++ SLG S + ED
Sbjct: 306 APEAQLLAMKVFTNSDTSATTGSSTLVSAIEDSAKIGADVLNMSLGSDSGNQTLEDPELA 365
Query: 281 GAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSV-----------------AASTIDR 323
+A E+G V GNSG V + + AS +
Sbjct: 366 AVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGTPGTSRGATTVASAENT 425
Query: 324 NFVSY---------LQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICK 374
+ ++ LQLG I + ++ TG ++K + +V + + D
Sbjct: 426 DVITQAVTITDGTGLQLGPGTIQLSSNDFTGSFDQKKFYVVKDASGNLSKGALADYTA-- 483
Query: 375 VGSLDPNKVKGKILFCLLREL---DGLVYAEEEAISGGSIGLVLGNDKQRGNDI--MAYA 429
KGKI EL D YA+ + G+ GL++ N+ + MA
Sbjct: 484 -------DAKGKIAIVKRGELSFDDKQKYAQ----AAGAAGLIIVNNDGTATPVTSMALT 532
Query: 430 HLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPII 489
PT ++ G+ + ++ A + A T V + P+ +
Sbjct: 533 TTFPTFGLSSVTGQKLVDWVTAHPDDSLGVKIALTLVPNQKYTEDKMSDFTSYGPVSNLS 592
Query: 490 LKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPN 549
KPDITAPG +I S Y SGTS++ P ++ ALLK N
Sbjct: 593 FKPDITAPGGNIW-------------STQNNNGYTNMSGTSMASPFIAGSQALLKQALNN 639
Query: 550 WSPAAF------KSAIMT--TTTIQGNNHRPIKDQSKED--ATPFGYGAGHIQPELAMD 598
+ + K +T T++ N +PI D + + +P GAG + + A+D
Sbjct: 640 KNNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLVDVKAAID 698
>P3P_LACLC (P15292) PIII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
Length = 1902
Score = 79.3 bits (194), Expect = 3e-14
Identities = 136/607 (22%), Positives = 231/607 (37%), Gaps = 113/607 (18%)
Query: 48 SYNKHINGFAAVLEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDS 107
SY +NGF+ + V + K+ + V +V K + ++ ++ +
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMANVQ---------A 199
Query: 108 IWEKGRY-GEGTIIANIDSGVSPESKSF--SDDGMGPVPSRWRGICQLDNFHCNRKLIGA 164
+W +Y GEGT+++ IDSG+ P K SDD + ++ F K G
Sbjct: 200 VWSNYKYKGEGTVVSVIDSGIDPTHKDMRLSDDKDVKLTKS-----DVEKFTDTVKH-GR 253
Query: 165 RFYSQ-GYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAK---GG 220
F S+ Y + N ++ + + HG + G N G + AK G
Sbjct: 254 YFNSKVPYGFNYADNNDTITDDKVDEQHGMHVAGIIGAN--------GTGDDPAKSVVGV 305
Query: 221 SPRSHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISI 280
+P + + A KV + A ++ A ED+ G D+++ SLG S + ED
Sbjct: 306 APEAQLLAMKVFSNSDTSAKTGSATVVSAIEDSAKIGADVLNMSLGSNSGNQTLEDPELA 365
Query: 281 GAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLFSV-----------------AASTIDR 323
+A E+G V GNSG V + + AS +
Sbjct: 366 AVQNANESGTAAVISAGNSGTSGSATEGVNKDYYGLQDNEMVGSPGTSRGATTVASAENT 425
Query: 324 NFVSY---------LQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICK 374
+ ++ LQLG + I + + TG ++K + +V + + D
Sbjct: 426 DVITQAVTITDGTGLQLGPETIQLSSHDFTGSFDQKKFYIVKDASGNLSKGALADYTA-- 483
Query: 375 VGSLDPNKVKGKILFCLLREL---DGLVYAEEEAISGGSIGLVLGNDKQRGNDI--MAYA 429
KGKI E D YA+ + G+ GL++ N + +A
Sbjct: 484 -------DAKGKIAIVKRGEFSFDDKQKYAQ----AAGAAGLIIVNTDGTATPMTSIALT 532
Query: 430 HLLPTSHINYTDGEYVHSYIKA-------TKTPMAYMTKAK-TEVGVKPAPVIASLSSRG 481
PT ++ G+ + ++ A K +A + K TE ++ +S G
Sbjct: 533 TTFPTFGLSSVTGQKLVDWVTAHPDDSLGVKITLAMLPNQKYTE------DKMSDFTSYG 586
Query: 482 PNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVA 541
P + + KPDITAPG +I S Y SGTS++ P ++ A
Sbjct: 587 P--VSNLSFKPDITAPGGNIW-------------STQNNNGYTNMSGTSMASPFIAGSQA 631
Query: 542 LLKTIYPNWSPAAF------KSAIMT--TTTIQGNNHRPIKDQSKED--ATPFGYGAGHI 591
LLK N + + K +T T++ N +PI D + + +P GAG +
Sbjct: 632 LLKQALNNKNNPFYAYYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLV 691
Query: 592 QPELAMD 598
+ A+D
Sbjct: 692 DVKAAID 698
>P2P_LACPA (Q02470) PII-type proteinase precursor (EC 3.4.21.96)
(Lactocepin) (Cell wall-associated serine proteinase)
(LP151)
Length = 1902
Score = 75.5 bits (184), Expect = 5e-13
Identities = 128/596 (21%), Positives = 222/596 (36%), Gaps = 91/596 (15%)
Query: 48 SYNKHINGFAAVLEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFLGLENNYGVVPKDS 107
SY +NGF+ + V + K+ + V +V K + ++ ++ +
Sbjct: 149 SYGYVVNGFSTKVRVVDIPKLKQIAGVKTVTLAKVYYPTDAKANSMANVQ---------A 199
Query: 108 IWEKGRY-GEGTIIANIDSGVSPESKSF--SDDGMGPVPSRWRGICQLDNFHCNRKLIGA 164
+W +Y GEGT+++ ID+G+ P K SDD + ++ F K G
Sbjct: 200 VWSNYKYKGEGTVVSVIDTGIDPTHKDMRLSDDKDVKLTKY-----DVEKFTDTAKH-GR 253
Query: 165 RFYSQ-GYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAK---GG 220
F S+ Y + N ++ + HG + G N G + K G
Sbjct: 254 YFTSKVPYGFNYADNNDTITDDTVDEQHGMHVAGIIGAN--------GTGDDPTKSVVGV 305
Query: 221 SPRSHVAAYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISI 280
+P + + A KV A ++ A ED+ G D+++ SLG S + ED
Sbjct: 306 APEAQLLAMKVFTNSDTSATTGSATLVSAIEDSAKIGADVLNMSLGSDSGNQTLEDPEIA 365
Query: 281 GAFHAIENGVIVVAGGGNSGPKFGTVTNVAPWLF-----------------SVAASTIDR 323
+A E+G V GNSG V + + AS +
Sbjct: 366 AVQNANESGTAAVISAGNSGTSGSATQGVNKDYYGLQDNEMVGTPGTSRGATTVASAENT 425
Query: 324 NFVSY---------LQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNATIEDAKICK 374
+ +S LQLG + I + ++ TG ++K + +V + D
Sbjct: 426 DVISQAVTITDGKDLQLGPETIQLSSNDFTGSFDQKKFYVVKDASGDLSKGAAADYTA-- 483
Query: 375 VGSLDPNKVKGKILFCLLRELDGLVYAEEEAISGGSIGLVLGNDKQRGNDI--MAYAHLL 432
KGKI EL+ ++ A + G+ GL++ N+ + +
Sbjct: 484 -------DAKGKIAIVKRGELN-FADKQKYAQAAGAAGLIIVNNDGTATPLTSIRLTTTF 535
Query: 433 PTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKP 492
PT ++ G+ + ++ A + A T + + P+ + KP
Sbjct: 536 PTFGLSSKTGQKLVDWVTAHPDDSLGVKIALTLLPNQKYTEDKMSDFTSYGPVSNLSFKP 595
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DITAPG +I S Y SGTS++ P ++ ALLK N +
Sbjct: 596 DITAPGGNIW-------------STQNNNGYTNMSGTSMASPFIAGSQALLKQALNNKNN 642
Query: 553 AAF------KSAIMT--TTTIQGNNHRPIKDQSKED--ATPFGYGAGHIQPELAMD 598
+ K +T T++ N +PI D + + +P GAG + + A+D
Sbjct: 643 PFYADYKQLKGTALTDFLKTVEMNTAQPINDINYNNVIVSPRRQGAGLVDVKAAID 698
>SUBF_BACSU (P16397) Bacillopeptidase F precursor (EC 3.4.21.-)
(Esterase) (RP-I protease) (90 kDa serine proteinase)
Length = 1433
Score = 51.2 bits (121), Expect = 1e-05
Identities = 43/135 (31%), Positives = 65/135 (47%), Gaps = 26/135 (19%)
Query: 455 PMAYMTKAKTEVGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGL 514
P ++ T A T++ K +A S +GP+P I KP+I+APGV+I S G
Sbjct: 396 PESFATGA-TDINKK----LADFSLQGPSPYDEI--KPEISAPGVNI------RSSVPGQ 442
Query: 515 ASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIK 574
++ W GTS++ PHVSA+ ALLK + S + + +T P+
Sbjct: 443 TYEDGW------DGTSMAGPHVSAVAALLKQANASLSVDEMEDILTSTA-------EPLT 489
Query: 575 DQSKEDATPFGYGAG 589
D + D+ GYG G
Sbjct: 490 DSTFPDSPNNGYGHG 504
Score = 35.8 bits (81), Expect = 0.42
Identities = 24/86 (27%), Positives = 37/86 (42%), Gaps = 11/86 (12%)
Query: 53 INGFAAVLEVEEAAKIAKHPNVVSVFENKGHELQTTRSWEFL-----------GLENNYG 101
+NG A E K+ + P V V N+ +L + S + G+E N
Sbjct: 145 VNGIAVHASKEVMEKVVQFPEVEKVLPNEKRQLFKSSSPFNMKKAQKAIKATDGVEWNVD 204
Query: 102 VVPKDSIWEKGRYGEGTIIANIDSGV 127
+ W G G GT++A+ID+GV
Sbjct: 205 QIDAPKAWALGYDGTGTVVASIDTGV 230
>SCA1_STRPY (P15926) C5a peptidase precursor (EC 3.4.21.-) (SCP)
Length = 1167
Score = 46.6 bits (109), Expect = 2e-04
Identities = 82/345 (23%), Positives = 139/345 (39%), Gaps = 62/345 (17%)
Query: 248 QAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAF-HAIENGVIVVAGGGNSGPKFGTV 306
QA DA++ G +I+ S G + + AF +A GV +V GN FG
Sbjct: 244 QAIRDAVNLGAKVINMSFGNAALAYANLPDETKKAFDYAKSKGVSIVTSAGNDS-SFGGK 302
Query: 307 TNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNAT 366
T + A D V D + ++++ P+++ L + K +
Sbjct: 303 TRLP------LADHPDYGVVGTPAAADSTL----TVASYSPDKQ---LTETAMVKTDDQQ 349
Query: 367 IEDAKICKVGSLDPNK------------------VKGKILFCLLRELDGLVYAEEEAISG 408
++ + +PNK VKGKI ++D A
Sbjct: 350 DKEMPVLSTNRFEPNKAYDYAYANRGMKEDDFKDVKGKIALIERGDID-FKDKVANAKKA 408
Query: 409 GSIGLVLGNDKQRGNDI-MAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTEVG 467
G++G+++ +++ +G I + +P + I+ DG + + T T A T G
Sbjct: 409 GAVGVLIYDNQDKGFPIELPNVDQMPAAFISRKDGLLLKDNPQKTITFNATPKVLPTASG 468
Query: 468 VKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGS 527
K ++ SS G I KPDI APG DIL + ++N++ S
Sbjct: 469 TK----LSRFSSWGLTADGNI--KPDIAAPGQDILSS----------VANNKYAKL---S 509
Query: 528 GTSISCPHVSAIVALL----KTIYPNWSPA----AFKSAIMTTTT 564
GTS+S P V+ I+ LL +T YP+ +P+ K +M++ T
Sbjct: 510 GTSMSAPLVAGIMGLLQKQYETQYPDMTPSERLDLAKKVLMSSAT 554
>SCA2_STRPY (P58099) C5a peptidase precursor (EC 3.4.21.-) (SCP)
Length = 1181
Score = 46.2 bits (108), Expect = 3e-04
Identities = 82/347 (23%), Positives = 142/347 (40%), Gaps = 66/347 (19%)
Query: 248 QAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAF-HAIENGVIVVAGGGNSGPKFGTV 306
QA DA++ G +I+ S G + + AF +A GV +V GN FG
Sbjct: 244 QAIIDAVNLGAKVINMSFGNAALAYANLPDETKKAFDYAKSKGVSIVTSAGNDS-SFGGK 302
Query: 307 TNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGLPNEKFYSLVSSVDAKVGNAT 366
T + A D V D + ++++ P+++ L + K +
Sbjct: 303 TRLP------LADHPDYGVVGTPAAADSTL----TVASYSPDKQ---LTETATVKTADQQ 349
Query: 367 IEDAKICKVGSLDPNK------------------VKGKILFCLLRELDGLVYAEE--EAI 406
++ + +PNK VKGKI ++D + ++ A
Sbjct: 350 DKEMPVLSTNRFEPNKAYDYAYANRGMKEDDFKDVKGKIALIERGDID---FKDKIANAK 406
Query: 407 SGGSIGLVLGNDKQRGNDI-MAYAHLLPTSHINYTDGEYVHSYIKATKTPMAYMTKAKTE 465
G++G+++ +++ +G I + +P + I+ DG + + T T A T
Sbjct: 407 KAGAVGVLIYDNQDKGFPIELPNVDQMPAAFISRKDGLLLKENPQKTITFNATPKVLPTA 466
Query: 466 VGVKPAPVIASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNI 525
G K ++ SS G I KPDI APG DIL + ++N++
Sbjct: 467 SGTK----LSRFSSWGLTADGNI--KPDIAAPGQDILSS----------VANNKYAKL-- 508
Query: 526 GSGTSISCPHVSAIVALL----KTIYPNWSPA----AFKSAIMTTTT 564
SGTS+S P V+ I+ LL +T YP+ +P+ K +M++ T
Sbjct: 509 -SGTSMSAPLVAGIMGLLQKQYETQYPDMTPSERLDLAKKVLMSSAT 554
>SUBT_BACAM (P00782) Subtilisin BPN' precursor (EC 3.4.21.62)
(Subtilisin Novo) (Subtilisin DFE) (Alkaline protease)
Length = 382
Score = 45.8 bits (107), Expect = 4e-04
Identities = 32/95 (33%), Positives = 48/95 (49%), Gaps = 21/95 (22%)
Query: 475 ASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCP 534
AS SS GP + D+ APGV I G N++ YN GTS++ P
Sbjct: 294 ASFSSVGP--------ELDVMAPGVSIQSTLPG----------NKYGAYN---GTSMASP 332
Query: 535 HVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNN 569
HV+ AL+ + +PNW+ +S++ TTT G++
Sbjct: 333 HVAGAAALILSKHPNWTNTQVRSSLENTTTKLGDS 367
Score = 40.8 bits (94), Expect = 0.013
Identities = 31/112 (27%), Positives = 52/112 (45%), Gaps = 10/112 (8%)
Query: 246 IMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGT 305
I+ E AI++ +D+I+ SLG S + + A+ +GV+VVA GN G +
Sbjct: 214 IINGIEWAIANNMDVINMSLGGPSGSAALKAAVD----KAVASGVVVVAAAGNEGTSGSS 269
Query: 306 VT----NVAPWLFSVAASTIDRNFVSYLQLGDKHIIM--GTSLSTGLPNEKF 351
T P + +V A S+ +G + +M G S+ + LP K+
Sbjct: 270 STVGYPGKYPSVIAVGAVDSSNQRASFSSVGPELDVMAPGVSIQSTLPGNKY 321
>TKSU_PYRKO (P58502) Tk-subtilisin precursor (EC 3.4.21.-)
Length = 422
Score = 45.4 bits (106), Expect = 5e-04
Identities = 56/232 (24%), Positives = 94/232 (40%), Gaps = 27/232 (11%)
Query: 176 GRLNQSLYNARDVLGHGTPTLSVAGG--------NFVSGANVFGLANGTAKGGSPRSHVA 227
G+++ L + D GHGT + G ++ + A+G S +A
Sbjct: 162 GKVSTKLRDCADQNGHGTHVIGTIAALNNDIGVVGVAPGVQIYSVRVLDARGSGSYSDIA 221
Query: 228 -AYKVCWLGTI*IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAI 286
+ LG + D D + A D D ++IS SLG + + D I I A++A
Sbjct: 222 IGIEQAILGPDGVADKDGDGIIA-GDPDDDAAEVISMSLGGPADDSYLYDMI-IQAYNA- 278
Query: 287 ENGVIVVAGGGNSGPKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIMGTSLSTGL 346
G+++VA GN G + P + +V A + N S+ + G + +
Sbjct: 279 --GIVIVAASGNEGAPSPSYPAAYPEVIAVGAIDSNDNIASFSNRQPEVSAPGVDILSTY 336
Query: 347 PNEKFYSL---------VSSVDAKVGNATIED-AKICKVGSLD---PNKVKG 385
P++ + +L VS V A + A + KI VG+ D N V+G
Sbjct: 337 PDDSYETLMGTSMATPHVSGVVALIQAAYYQKYGKILPVGTFDDISKNTVRG 388
Score = 44.3 bits (103), Expect = 0.001
Identities = 41/132 (31%), Positives = 59/132 (44%), Gaps = 29/132 (21%)
Query: 474 IASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISC 533
IAS S+R +P+++APGVDIL Y D+ Y GTS++
Sbjct: 315 IASFSNR----------QPEVSAPGVDILSTY----------PDDS---YETLMGTSMAT 351
Query: 534 PHVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFG----YGAG 589
PHVS +VAL++ Y + + T I N R I + +D P G YG G
Sbjct: 352 PHVSGVVALIQAAY--YQKYGKILPVGTFDDISKNTVRGILHITADDLGPTGWDADYGYG 409
Query: 590 HIQPELAMDPGL 601
++ LA+ L
Sbjct: 410 VVRAALAVQAAL 421
>PRTM_BACSK (Q99405) M-protease (EC 3.4.21.-)
Length = 269
Score = 44.3 bits (103), Expect = 0.001
Identities = 25/78 (32%), Positives = 39/78 (49%), Gaps = 13/78 (16%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV++ Y G+ Y +GTS++ PHV+ + AL+K P+WS
Sbjct: 191 DIVAPGVNVQSTYPGST-------------YASLNGTSMATPHVAGVAALVKQKNPSWSN 237
Query: 553 AAFKSAIMTTTTIQGNNH 570
++ + T T GN +
Sbjct: 238 VQIRNHLKNTATGLGNTN 255
Score = 42.4 bits (98), Expect = 0.004
Identities = 47/175 (26%), Positives = 77/175 (43%), Gaps = 22/175 (12%)
Query: 184 NARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTD 243
+ +D GHGT G + N G+ G +P + + A KV +
Sbjct: 55 STQDGNGHGTHV----AGTIAALNNSIGVL-----GVAPSAELYAVKVLGASG---SGSV 102
Query: 244 ADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKF 303
+ I Q E A ++G+ + + SLG SP E ++ A GV+VVA GNSG
Sbjct: 103 SSIAQGLEWAGNNGMHVANLSLGSPSPSATLEQAVN----SATSRGVLVVAASGNSGA-- 156
Query: 304 GTVTNVAPWLFSVAASTIDR--NFVSYLQLGDKHIIM--GTSLSTGLPNEKFYSL 354
G+++ A + ++A D+ N S+ Q G I+ G ++ + P + SL
Sbjct: 157 GSISYPARYANAMAVGATDQNNNRASFSQYGAGLDIVAPGVNVQSTYPGSTYASL 211
>ELYA_BACHD (P41363) Thermostable alkaline protease precursor (EC
3.4.21.-)
Length = 361
Score = 43.5 bits (101), Expect = 0.002
Identities = 29/94 (30%), Positives = 49/94 (51%), Gaps = 21/94 (22%)
Query: 475 ASLSSRGPNPIQPIILKPDITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCP 534
AS S+ GP + +I+APGV++ Y G N+++ SGTS++ P
Sbjct: 273 ASFSTYGP--------EIEISAPGVNVNSTYTG----------NRYVSL---SGTSMATP 311
Query: 535 HVSAIVALLKTIYPNWSPAAFKSAIMTTTTIQGN 568
HV+ + AL+K+ YP+++ + I T T G+
Sbjct: 312 HVAGVAALVKSRYPSYTNNQIRQRINQTATYLGS 345
>SUBD_BACLI (P00781) Subtilisin DY (EC 3.4.21.62)
Length = 274
Score = 42.7 bits (99), Expect = 0.003
Identities = 52/210 (24%), Positives = 92/210 (43%), Gaps = 25/210 (11%)
Query: 198 VAGGNFVSGA--NVFGLANGTAKGGSPRS------------HVAAYKVCWLGTI*IECTD 243
V G +FVSG N G +GT G+ + +V+ Y + L + T
Sbjct: 45 VGGASFVSGESYNTDGNGHGTHVAGTVAALDNTTGVLGVAPNVSLYAIKVLNSSG-SGTY 103
Query: 244 ADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGP-- 301
+ I+ E A +G+D+I+ SLG S + + A +G++VVA GNSG
Sbjct: 104 SAIVSGIEWATQNGLDVINMSLGGPSGSTALKQAVD----KAYASGIVVVAAAGNSGSSG 159
Query: 302 KFGTVTNVAPWLFSVAASTID--RNFVSYLQLGDKHIIM--GTSLSTGLPNEKFYSLVSS 357
T+ A + +A +D +N S+ +G + +M G S+ + P+ + SL +
Sbjct: 160 SQNTIGYPAKYDSVIAVGAVDSNKNRASFSSVGAELEVMAPGVSVYSTYPSNTYTSLNGT 219
Query: 358 VDAKVGNATIEDAKICKVGSLDPNKVKGKI 387
A A + K +L ++V+ ++
Sbjct: 220 SMASPHVAGAAALILSKYPTLSASQVRNRL 249
Score = 35.8 bits (81), Expect = 0.42
Identities = 28/104 (26%), Positives = 44/104 (41%), Gaps = 27/104 (25%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
++ APGV + Y N + N GTS++ PHV+ AL+ + YP S
Sbjct: 196 EVMAPGVSVYSTY----------PSNTYTSLN---GTSMASPHVAGAAALILSKYPTLSA 242
Query: 553 AAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELA 596
+ ++ + +T T G++ F YG G I E A
Sbjct: 243 SQVRNRLSSTATNLGDS--------------FYYGKGLINVEAA 272
>ELYA_BACAO (P27693) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 42.7 bits (99), Expect = 0.003
Identities = 47/175 (26%), Positives = 77/175 (43%), Gaps = 22/175 (12%)
Query: 184 NARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTD 243
+ +D GHGT G + N G+ G +P + + A KV +
Sbjct: 166 STQDGNGHGTHV----AGTIAALNNSIGVL-----GVAPNAELYAVKVLGASG---SGSV 213
Query: 244 ADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKF 303
+ I Q E A ++G+ + + SLG SP E ++ A GV+VVA GNSG
Sbjct: 214 SSIAQGLEWAGNNGMHVANLSLGSPSPSATLEQAVN----SATSRGVLVVAASGNSGA-- 267
Query: 304 GTVTNVAPWLFSVAASTIDR--NFVSYLQLGDKHIIM--GTSLSTGLPNEKFYSL 354
G+++ A + ++A D+ N S+ Q G I+ G ++ + P + SL
Sbjct: 268 GSISYPARYANAMAVGATDQNNNRASFSQYGAGLDIVAPGVNVQSTYPGSTYASL 322
Score = 41.6 bits (96), Expect = 0.008
Identities = 24/78 (30%), Positives = 38/78 (47%), Gaps = 13/78 (16%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV++ Y G+ Y +GTS++ PHV+ AL+K P+WS
Sbjct: 302 DIVAPGVNVQSTYPGST-------------YASLNGTSMATPHVAGAAALVKQKNPSWSN 348
Query: 553 AAFKSAIMTTTTIQGNNH 570
++ + T T G+ +
Sbjct: 349 VQIRNHLKNTATSLGSTN 366
>SUBS_BACLE (P29600) Subtilisin Savinase (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 42.4 bits (98), Expect = 0.004
Identities = 47/175 (26%), Positives = 77/175 (43%), Gaps = 22/175 (12%)
Query: 184 NARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTD 243
+ +D GHGT G + N G+ G +P + + A KV +
Sbjct: 55 STQDGNGHGTHV----AGTIAALNNSIGVL-----GVAPSAELYAVKVLGASG---SGSV 102
Query: 244 ADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKF 303
+ I Q E A ++G+ + + SLG SP E ++ A GV+VVA GNSG
Sbjct: 103 SSIAQGLEWAGNNGMHVANLSLGSPSPSATLEQAVN----SATSRGVLVVAASGNSGA-- 156
Query: 304 GTVTNVAPWLFSVAASTIDR--NFVSYLQLGDKHIIM--GTSLSTGLPNEKFYSL 354
G+++ A + ++A D+ N S+ Q G I+ G ++ + P + SL
Sbjct: 157 GSISYPARYANAMAVGATDQNNNRASFSQYGAGLDIVAPGVNVQSTYPGSTYASL 211
Score = 41.6 bits (96), Expect = 0.008
Identities = 24/78 (30%), Positives = 38/78 (47%), Gaps = 13/78 (16%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV++ Y G+ Y +GTS++ PHV+ AL+K P+WS
Sbjct: 191 DIVAPGVNVQSTYPGST-------------YASLNGTSMATPHVAGAAALVKQKNPSWSN 237
Query: 553 AAFKSAIMTTTTIQGNNH 570
++ + T T G+ +
Sbjct: 238 VQIRNHLKNTATSLGSTN 255
>ELYA_BACYA (P20724) Alkaline elastase YaB precursor (EC 3.4.21.-)
Length = 378
Score = 42.4 bits (98), Expect = 0.004
Identities = 27/76 (35%), Positives = 37/76 (48%), Gaps = 13/76 (17%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV + G G AS N GTS++ PHV+ + AL+K P+WS
Sbjct: 300 DIVAPGVGVQSTVPG----NGYASFN---------GTSMATPHVAGVAALVKQKNPSWSN 346
Query: 553 AAFKSAIMTTTTIQGN 568
++ + T T GN
Sbjct: 347 VQIRNHLKNTATNLGN 362
Score = 37.0 bits (84), Expect = 0.19
Identities = 36/133 (27%), Positives = 57/133 (42%), Gaps = 7/133 (5%)
Query: 192 GTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTDADIMQAFE 251
G P +S G+ A N + +V Y V LG + + I Q +
Sbjct: 161 GEPNISDGNGHGTQVAGTIAALNNSIGVLGVAPNVDLYGVKVLGASG-SGSISGIAQGLQ 219
Query: 252 DAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKFGTVTNVAP 311
A ++G+ I + SLG ++ E ++ A +GV+VVA GNSG G V A
Sbjct: 220 WAANNGMHIANMSLGSSAGSATMEQAVN----QATASGVLVVAASGNSGA--GNVGFPAR 273
Query: 312 WLFSVAASTIDRN 324
+ ++A D+N
Sbjct: 274 YANAMAVGATDQN 286
>ELYA_BACCS (P41362) Alkaline protease precursor (EC 3.4.21.-)
Length = 380
Score = 42.4 bits (98), Expect = 0.004
Identities = 47/175 (26%), Positives = 77/175 (43%), Gaps = 22/175 (12%)
Query: 184 NARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWLGTI*IECTD 243
+ +D GHGT G + N G+ G +P + + A KV +
Sbjct: 166 STQDGNGHGTHV----AGTIAALNNSIGVL-----GVAPSAELYAVKVLGASG---SGSV 213
Query: 244 ADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSGPKF 303
+ I Q E A ++G+ + + SLG SP E ++ A GV+VVA GNSG
Sbjct: 214 SSIAQGLEWAGNNGMHVANLSLGSPSPSATLEQAVN----SATSRGVLVVAASGNSGA-- 267
Query: 304 GTVTNVAPWLFSVAASTIDR--NFVSYLQLGDKHIIM--GTSLSTGLPNEKFYSL 354
G+++ A + ++A D+ N S+ Q G I+ G ++ + P + SL
Sbjct: 268 GSISYPARYANAMAVGATDQNNNRASFSQYGAGLDIVAPGVNVQSTYPGSTYASL 322
Score = 41.6 bits (96), Expect = 0.008
Identities = 24/78 (30%), Positives = 38/78 (47%), Gaps = 13/78 (16%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV++ Y G+ Y +GTS++ PHV+ AL+K P+WS
Sbjct: 302 DIVAPGVNVQSTYPGST-------------YASLNGTSMATPHVAGAAALVKQKNPSWSN 348
Query: 553 AAFKSAIMTTTTIQGNNH 570
++ + T T G+ +
Sbjct: 349 VQIRNHLKNTATSLGSTN 366
>SUBT_BACLI (P00780) Subtilisin Carlsberg precursor (EC 3.4.21.62)
Length = 379
Score = 42.0 bits (97), Expect = 0.006
Identities = 55/215 (25%), Positives = 93/215 (42%), Gaps = 26/215 (12%)
Query: 194 PTLSVAGG-NFVSGA--NVFGLANGTAKGGSPRS------------HVAAYKVCWLGTI* 238
P L+V GG +FV+G N G +GT G+ + V+ Y V L +
Sbjct: 145 PDLNVVGGASFVAGEAYNTDGNGHGTHVAGTVAALDNTTGVLGVAPSVSLYAVKVLNSSG 204
Query: 239 IECTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGN 298
T + I+ E A ++G+D+I+ SLG S + + +A GV+VVA GN
Sbjct: 205 -SGTYSGIVSGIEWATTNGMDVINMSLGGPSGSTAMKQAVD----NAYARGVVVVAAAGN 259
Query: 299 SGPKFGTVTNVAPWLFS--VAASTIDRNF--VSYLQLGDKHIIM--GTSLSTGLPNEKFY 352
SG T T P + +A +D N S+ +G + +M G + + P +
Sbjct: 260 SGSSGNTNTIGYPAKYDSVIAVGAVDSNSNRASFSSVGAELEVMAPGAGVYSTYPTSTYA 319
Query: 353 SLVSSVDAKVGNATIEDAKICKVGSLDPNKVKGKI 387
+L + A A + K +L ++V+ ++
Sbjct: 320 TLNGTSMASPHVAGAAALILSKHPNLSASQVRNRL 354
Score = 36.2 bits (82), Expect = 0.32
Identities = 28/104 (26%), Positives = 45/104 (42%), Gaps = 27/104 (25%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
++ APG + Y PT Y +GTS++ PHV+ AL+ + +PN S
Sbjct: 301 EVMAPGAGVYSTY-----PTST--------YATLNGTSMASPHVAGAAALILSKHPNLSA 347
Query: 553 AAFKSAIMTTTTIQGNNHRPIKDQSKEDATPFGYGAGHIQPELA 596
+ ++ + +T T G++ F YG G I E A
Sbjct: 348 SQVRNRLSSTATYLGSS--------------FYYGKGLINVEAA 377
>SUBB_BACLE (P29599) Subtilisin BL (EC 3.4.21.62) (Alkaline
protease)
Length = 269
Score = 41.6 bits (96), Expect = 0.008
Identities = 24/78 (30%), Positives = 38/78 (47%), Gaps = 13/78 (16%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
DI APGV++ Y G+ Y +GTS++ PHV+ AL+K P+WS
Sbjct: 191 DIVAPGVNVQSTYPGST-------------YASLNGTSMATPHVAGAAALVKQKNPSWSN 237
Query: 553 AAFKSAIMTTTTIQGNNH 570
++ + T T G+ +
Sbjct: 238 VQIRNHLKNTATSLGSTN 255
Score = 40.0 bits (92), Expect = 0.022
Identities = 47/176 (26%), Positives = 73/176 (40%), Gaps = 24/176 (13%)
Query: 184 NARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVAAYKVCWL---GTI*IE 240
+ +D GHGT G + N G+ G +P + + A KV G I
Sbjct: 55 STQDGNGHGTHV----AGTIAALNNSIGVL-----GVAPSAELYAVKVLGADGRGAI--- 102
Query: 241 CTDADIMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAFHAIENGVIVVAGGGNSG 300
+ I Q E A ++G+ + + SLG SP E ++ A GV+VVA GNSG
Sbjct: 103 ---SSIAQGLEWAGNNGMHVANLSLGSPSPSATLEQAVN----SATSRGVLVVAASGNSG 155
Query: 301 PKFGTVTNVAPWLFSVAASTIDRNFVSYLQLGDKHIIM--GTSLSTGLPNEKFYSL 354
+ +V A+ + N S+ Q G I+ G ++ + P + SL
Sbjct: 156 ASSISYPARYANAMAVGATDQNNNRASFSQYGAGLDIVAPGVNVQSTYPGSTYASL 211
>SUBT_BACSU (P04189) Subtilisin E precursor (EC 3.4.21.62)
Length = 381
Score = 40.8 bits (94), Expect = 0.013
Identities = 23/77 (29%), Positives = 36/77 (45%), Gaps = 13/77 (16%)
Query: 493 DITAPGVDILYAYIGAISPTGLASDNQWIPYNIGSGTSISCPHVSAIVALLKTIYPNWSP 552
D+ APGV I G Y +GTS++ PHV+ AL+ + +P W+
Sbjct: 303 DVMAPGVSIQSTLPGGT-------------YGAYNGTSMATPHVAGAAALILSKHPTWTN 349
Query: 553 AAFKSAIMTTTTIQGNN 569
A + + +T T GN+
Sbjct: 350 AQVRDRLESTATYLGNS 366
Score = 38.9 bits (89), Expect = 0.050
Identities = 67/310 (21%), Positives = 115/310 (36%), Gaps = 77/310 (24%)
Query: 51 KHINGFAAVLEVEEAAKIAKHPNVVSVFENK-GHELQTTRSWEFLGLENNYGV--VPKDS 107
K++N AA L+ + ++ K P+V V E+ HE + YG+ + +
Sbjct: 71 KYVNAAAATLDEKAVKELKKDPSVAYVEEDHIAHEYAQSVP---------YGISQIKAPA 121
Query: 108 IWEKGRYGEGTIIANIDSGVSPESKSFSDDGMGPVPSRWRGICQLDNFHCNRKLIGARFY 167
+ +G G +A IDSG+ D+ H + + G +
Sbjct: 122 LHSQGYTGSNVKVAVIDSGI-------------------------DSSHPDLNVRGGASF 156
Query: 168 SQGYESKFGRLNQSLYNARDVLGHGTPTLSVAGGNFVSGANVFGLANGTAKGGSPRSHVA 227
+ + +D HGT G + N G+ G SP + +
Sbjct: 157 VPSETNPY----------QDGSSHGTHV----AGTIAALNNSIGVL-----GVSPSASLY 197
Query: 228 AYKVCWLGTI*IECTDAD----IMQAFEDAISDGVDIISCSLGQTSPKEFFEDGISIGAF 283
A KV ++ T + I+ E AIS+ +D+I+ SLG + + +
Sbjct: 198 AVKV-------LDSTGSGQYSWIINGIEWAISNNMDVINMSLGGPTGSTALKTVVD---- 246
Query: 284 HAIENGVIVVAGGGNSGPKFGTVT----NVAPWLFSVAASTIDRNFVSYLQLGDKHIIM- 338
A+ +G++V A GN G T T P +V A S+ G + +M
Sbjct: 247 KAVSSGIVVAAAAGNEGSSGSTSTVGYPAKYPSTIAVGAVNSSNQRASFSSAGSELDVMA 306
Query: 339 -GTSLSTGLP 347
G S+ + LP
Sbjct: 307 PGVSIQSTLP 316
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 94,970,549
Number of Sequences: 164201
Number of extensions: 4351759
Number of successful extensions: 9645
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 9510
Number of HSP's gapped (non-prelim): 127
length of query: 740
length of database: 59,974,054
effective HSP length: 118
effective length of query: 622
effective length of database: 40,598,336
effective search space: 25252164992
effective search space used: 25252164992
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC147712.15


