

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147537.6 + phase: 0
(80 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
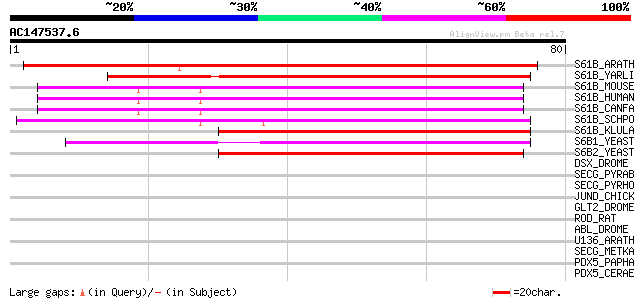
Score E
Sequences producing significant alignments: (bits) Value
S61B_ARATH (P38389) Protein transport protein SEC61 beta subunit 130 5e-31
S61B_YARLI (Q9HFC7) Protein transport protein SEC61 beta subunit 60 1e-09
S61B_MOUSE (Q9CQS8) Protein transport protein SEC61 beta subunit 58 4e-09
S61B_HUMAN (P60468) Protein transport protein SEC61 beta subunit 58 4e-09
S61B_CANFA (P60467) Protein transport protein SEC61 beta subunit 58 4e-09
S61B_SCHPO (O43002) Prpbable protein transport protein SEC61 bet... 56 1e-08
S61B_KLULA (Q8J2P4) Protein transport protein SEC61 beta subunit 49 3e-06
S6B1_YEAST (P52870) Protein transport protein SEC61 beta 1 subunit 45 3e-05
S6B2_YEAST (P52871) Protein transport protein SEC61 beta 2 subunit 44 7e-05
DSX_DROME (P23023) Doublesex protein 34 0.059
SECG_PYRAB (P60462) Preprotein translocase secG subunit (Protein... 32 0.22
SECG_PYRHO (P60463) Preprotein translocase secG subunit (Protein... 31 0.65
JUND_CHICK (P27921) Transcription factor jun-D 31 0.65
GLT2_DROME (Q6WV19) Polypeptide N-acetylgalactosaminyltransferas... 31 0.65
ROD_RAT (Q9JJ54) Heterogeneous nuclear ribonucleoprotein D0 (hnR... 30 1.1
ABL_DROME (P00522) Tyrosine-protein kinase Abl (EC 2.7.1.112) (D... 30 1.1
U136_ARATH (O64847) UPF0136 protein At2g26240 30 1.5
SECG_METKA (Q8TYT4) Preprotein translocase secG subunit (Protein... 30 1.5
PDX5_PAPHA (Q9GLW9) Peroxiredoxin 5, mitochondrial precursor (Pr... 30 1.5
PDX5_CERAE (Q9GLW7) Peroxiredoxin 5, mitochondrial precursor (Pr... 30 1.5
>S61B_ARATH (P38389) Protein transport protein SEC61 beta subunit
Length = 82
Score = 130 bits (328), Expect = 5e-31
Identities = 67/78 (85%), Positives = 71/78 (90%), Gaps = 4/78 (5%)
Query: 3 LGGGAPQRGSAAATASMRRRK----TAGGGASGGAAGTMLQFYTDDAPGLKISPNVVLVM 58
+G GAPQRGSAAATASMRRRK GGGASGGAAG+MLQFYTDDAPGLKISPNVVL+M
Sbjct: 2 VGSGAPQRGSAAATASMRRRKPTSGAGGGGASGGAAGSMLQFYTDDAPGLKISPNVVLIM 61
Query: 59 SIGFIAFVAVLHVVGKLY 76
SIGFIAFVAVLHV+GKLY
Sbjct: 62 SIGFIAFVAVLHVMGKLY 79
>S61B_YARLI (Q9HFC7) Protein transport protein SEC61 beta subunit
Length = 91
Score = 59.7 bits (143), Expect = 1e-09
Identities = 30/61 (49%), Positives = 42/61 (68%), Gaps = 1/61 (1%)
Query: 15 ATASMRRRKTAGGGASGGAAGTMLQFYTDDAPGLKISPNVVLVMSIGFIAFVAVLHVVGK 74
A AS R T GA GG++ TML+ YTD++ GLK+ P VV+V+S+GFI V LH++ K
Sbjct: 26 AKASQRPTSTRSVGA-GGSSSTMLKLYTDESQGLKVDPVVVMVLSLGFIFSVVALHILAK 84
Query: 75 L 75
+
Sbjct: 85 V 85
>S61B_MOUSE (Q9CQS8) Protein transport protein SEC61 beta subunit
Length = 95
Score = 58.2 bits (139), Expect = 4e-09
Identities = 34/78 (43%), Positives = 47/78 (59%), Gaps = 8/78 (10%)
Query: 5 GGAPQRGSAAATA--SMRRRKTAG------GGASGGAAGTMLQFYTDDAPGLKISPNVVL 56
G +P + AA A ++R+RK A G + G M +FYT+D+PGLK+ P VL
Sbjct: 14 GRSPSKAVAARAAGSTVRQRKNASCGTRSAGRTTSAGTGGMWRFYTEDSPGLKVGPVPVL 73
Query: 57 VMSIGFIAFVAVLHVVGK 74
VMS+ FIA V +LH+ GK
Sbjct: 74 VMSLLFIAAVFMLHIWGK 91
>S61B_HUMAN (P60468) Protein transport protein SEC61 beta subunit
Length = 95
Score = 58.2 bits (139), Expect = 4e-09
Identities = 34/78 (43%), Positives = 47/78 (59%), Gaps = 8/78 (10%)
Query: 5 GGAPQRGSAAATA--SMRRRKTAG------GGASGGAAGTMLQFYTDDAPGLKISPNVVL 56
G +P + AA A ++R+RK A G + G M +FYT+D+PGLK+ P VL
Sbjct: 14 GRSPSKAVAARAAGSTVRQRKNASCGTRSAGRTTSAGTGGMWRFYTEDSPGLKVGPVPVL 73
Query: 57 VMSIGFIAFVAVLHVVGK 74
VMS+ FIA V +LH+ GK
Sbjct: 74 VMSLLFIASVFMLHIWGK 91
>S61B_CANFA (P60467) Protein transport protein SEC61 beta subunit
Length = 95
Score = 58.2 bits (139), Expect = 4e-09
Identities = 34/78 (43%), Positives = 47/78 (59%), Gaps = 8/78 (10%)
Query: 5 GGAPQRGSAAATA--SMRRRKTAG------GGASGGAAGTMLQFYTDDAPGLKISPNVVL 56
G +P + AA A ++R+RK A G + G M +FYT+D+PGLK+ P VL
Sbjct: 14 GRSPSKAVAARAAGSTVRQRKNASCGTRSAGRTTSAGTGGMWRFYTEDSPGLKVGPVPVL 73
Query: 57 VMSIGFIAFVAVLHVVGK 74
VMS+ FIA V +LH+ GK
Sbjct: 74 VMSLLFIASVFMLHIWGK 91
>S61B_SCHPO (O43002) Prpbable protein transport protein SEC61 beta
subunit
Length = 102
Score = 56.2 bits (134), Expect = 1e-08
Identities = 32/79 (40%), Positives = 46/79 (57%), Gaps = 5/79 (6%)
Query: 2 ALGGGAPQRGSAAATASMRRRKTAG--GGASGGAAG---TMLQFYTDDAPGLKISPNVVL 56
A GG Q AA + +G G + GA G T+L+ YTD+A G K+ P VV+
Sbjct: 17 APGGPKSQIRRRAAVEKNTKESNSGPAGARAAGAPGSTPTLLKLYTDEASGFKVDPVVVM 76
Query: 57 VMSIGFIAFVAVLHVVGKL 75
V+S+GFIA V +LH+V ++
Sbjct: 77 VLSVGFIASVFLLHIVARI 95
>S61B_KLULA (Q8J2P4) Protein transport protein SEC61 beta subunit
Length = 88
Score = 48.5 bits (114), Expect = 3e-06
Identities = 20/45 (44%), Positives = 33/45 (72%)
Query: 31 GGAAGTMLQFYTDDAPGLKISPNVVLVMSIGFIAFVAVLHVVGKL 75
GG++ ++L+ YTD+A GL++ P VVL +++ F+ V LHVV K+
Sbjct: 39 GGSSSSILKLYTDEANGLRVDPLVVLFLAVAFVFSVVALHVVAKV 83
>S6B1_YEAST (P52870) Protein transport protein SEC61 beta 1
subunit
Length = 82
Score = 45.4 bits (106), Expect = 3e-05
Identities = 23/67 (34%), Positives = 39/67 (57%), Gaps = 6/67 (8%)
Query: 9 QRGSAAATASMRRRKTAGGGASGGAAGTMLQFYTDDAPGLKISPNVVLVMSIGFIAFVAV 68
++GS+ A+ +K S +L+ Y+D+A GL++ P VVL +++GFI V
Sbjct: 17 KQGSSQKVAASAPKKNTNSNNS------ILKIYSDEATGLRVDPLVVLFLAVGFIFSVVA 70
Query: 69 LHVVGKL 75
LHV+ K+
Sbjct: 71 LHVISKV 77
>S6B2_YEAST (P52871) Protein transport protein SEC61 beta 2
subunit
Length = 88
Score = 43.9 bits (102), Expect = 7e-05
Identities = 19/44 (43%), Positives = 31/44 (70%)
Query: 31 GGAAGTMLQFYTDDAPGLKISPNVVLVMSIGFIAFVAVLHVVGK 74
GG++ ++L+ YTD+A G ++ VVL +S+GFI V LH++ K
Sbjct: 40 GGSSSSILKLYTDEANGFRVDSLVVLFLSVGFIFSVIALHLLTK 83
>DSX_DROME (P23023) Doublesex protein
Length = 549
Score = 34.3 bits (77), Expect = 0.059
Identities = 15/32 (46%), Positives = 18/32 (55%)
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGGAAG 35
GGGAP +AAA S GGG GG++G
Sbjct: 253 GGGAPSSSNAAAATSSNGSSGGGGGGGGGSSG 284
>SECG_PYRAB (P60462) Preprotein translocase secG subunit (Protein
transport protein SEC61 beta subunit homolog)
Length = 56
Score = 32.3 bits (72), Expect = 0.22
Identities = 12/37 (32%), Positives = 24/37 (64%)
Query: 37 MLQFYTDDAPGLKISPNVVLVMSIGFIAFVAVLHVVG 73
+++F+ +D +KI+P + +++ I F +LHVVG
Sbjct: 15 LMRFFDEDTRAIKITPKGAVALTLILIIFEIILHVVG 51
>SECG_PYRHO (P60463) Preprotein translocase secG subunit (Protein
transport protein SEC61 beta subunit homolog)
Length = 56
Score = 30.8 bits (68), Expect = 0.65
Identities = 12/37 (32%), Positives = 23/37 (61%)
Query: 37 MLQFYTDDAPGLKISPNVVLVMSIGFIAFVAVLHVVG 73
+++F+ +D +KI+P + + + I F +LHVVG
Sbjct: 15 LMRFFDEDTRAIKITPKGAIALVLILIIFEILLHVVG 51
>JUND_CHICK (P27921) Transcription factor jun-D
Length = 323
Score = 30.8 bits (68), Expect = 0.65
Identities = 22/56 (39%), Positives = 24/56 (42%), Gaps = 14/56 (25%)
Query: 4 GGGAPQRGSAAATASMRRRKTAGGGASGGAAGTMLQFYTDDAPGLKISPNVVLVMS 59
GGG P G+AAA GGG GG G L APGL P V +S
Sbjct: 143 GGGGPNGGAAAA---------GGGGGGGGGGGGEL-----PAPGLAPEPPVYANLS 184
>GLT2_DROME (Q6WV19) Polypeptide N-acetylgalactosaminyltransferase
2 (EC 2.4.1.41) (Protein-UDP
acetylgalactosaminyltransferase 2)
(UDP-GalNAc:polypeptide
N-acetylgalactosaminyltransferase 2) (pp-GaNTase 2)
Length = 633
Score = 30.8 bits (68), Expect = 0.65
Identities = 20/49 (40%), Positives = 26/49 (52%), Gaps = 5/49 (10%)
Query: 12 SAAATASMRRRKTAGGGASGGAAGTMLQFYTDDAPGLKISPNVVLVMSI 60
SAAA ++ R+ AGGGA GAA DD G NV+L+ S+
Sbjct: 53 SAAAASTARQWAPAGGGAGPGAAAGAAGSGADDPGG-----NVILIGSV 96
>ROD_RAT (Q9JJ54) Heterogeneous nuclear ribonucleoprotein D0
(hnRNP D0) (AU-rich element RNA-binding protein 1)
Length = 353
Score = 30.0 bits (66), Expect = 1.1
Identities = 16/49 (32%), Positives = 25/49 (50%)
Query: 2 ALGGGAPQRGSAAATASMRRRKTAGGGASGGAAGTMLQFYTDDAPGLKI 50
A+GG A ++ A A+ AG G+ GG+A + + +A G KI
Sbjct: 18 AVGGSAGEQEGAMVAAAQGAAAAAGSGSGGGSAAGGTEGGSTEAEGAKI 66
>ABL_DROME (P00522) Tyrosine-protein kinase Abl (EC 2.7.1.112)
(D-ash)
Length = 1520
Score = 30.0 bits (66), Expect = 1.1
Identities = 17/54 (31%), Positives = 26/54 (47%)
Query: 3 LGGGAPQRGSAAATASMRRRKTAGGGASGGAAGTMLQFYTDDAPGLKISPNVVL 56
LGGG+ RGS ++S +G G GG +G+ L + + +P V L
Sbjct: 51 LGGGSTLRGSRIKSSSSGVASGSGSGGGGGGSGSGLSQRSGGHKDARCNPTVGL 104
>U136_ARATH (O64847) UPF0136 protein At2g26240
Length = 108
Score = 29.6 bits (65), Expect = 1.5
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 6/50 (12%)
Query: 29 ASGGAAGTMLQFYTDDAPGLKISPNVVLVMSIGFIAFVAVLHVVGKLYLR 78
A GG+A YT+ PG N VL SIG + A+ ++G YLR
Sbjct: 36 AGGGSAALFYYVYTE-LPG-----NPVLASSIGIVGSAALTGMMGSRYLR 79
>SECG_METKA (Q8TYT4) Preprotein translocase secG subunit (Protein
transport protein SEC61 beta subunit homolog)
Length = 57
Score = 29.6 bits (65), Expect = 1.5
Identities = 11/40 (27%), Positives = 21/40 (52%)
Query: 33 AAGTMLQFYTDDAPGLKISPNVVLVMSIGFIAFVAVLHVV 72
A +L F+ ++APG+KI P+ +L + + H +
Sbjct: 15 ARAGILSFWDEEAPGIKIDPDYILYACFAVAVLLIIAHTM 54
>PDX5_PAPHA (Q9GLW9) Peroxiredoxin 5, mitochondrial precursor
(Prx-V)
Length = 215
Score = 29.6 bits (65), Expect = 1.5
Identities = 24/49 (48%), Positives = 26/49 (52%), Gaps = 9/49 (18%)
Query: 3 LGGGAPQRGSAAATASMRRRKTAGGGASGG------AAGTMLQFYTDDA 45
LGG A Q S AATA+ RRR + GG ASGG AA M DA
Sbjct: 17 LGGAAGQ--SVAATAAARRR-SEGGWASGGVRSFSRAAAAMAPIKVGDA 62
>PDX5_CERAE (Q9GLW7) Peroxiredoxin 5, mitochondrial precursor
(Prx-V)
Length = 215
Score = 29.6 bits (65), Expect = 1.5
Identities = 24/49 (48%), Positives = 26/49 (52%), Gaps = 9/49 (18%)
Query: 3 LGGGAPQRGSAAATASMRRRKTAGGGASGG------AAGTMLQFYTDDA 45
LGG A Q S AATA+ RRR + GG ASGG AA M DA
Sbjct: 17 LGGAARQ--SVAATAAARRR-SEGGWASGGVRSFSRAAAAMAPIKVGDA 62
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.136 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,196,610
Number of Sequences: 164201
Number of extensions: 331553
Number of successful extensions: 2874
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 2600
Number of HSP's gapped (non-prelim): 257
length of query: 80
length of database: 59,974,054
effective HSP length: 56
effective length of query: 24
effective length of database: 50,778,798
effective search space: 1218691152
effective search space used: 1218691152
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147537.6


