

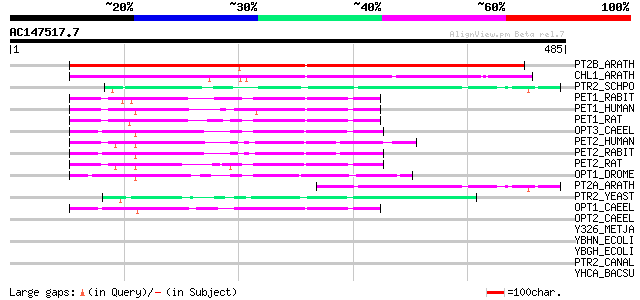
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.7 - phase: 0
(485 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
PT2B_ARATH (P46032) Peptide transporter PTR2-B (Histidine transp... 323 6e-88
CHL1_ARATH (Q05085) Nitrate/chlorate transporter 285 1e-76
PTR2_SCHPO (Q9P380) Probable peptide transporter Ptr2 (Peptide p... 76 2e-13
PET1_RABIT (P36836) Oligopeptide transporter, small intestine is... 70 2e-11
PET1_HUMAN (P46059) Oligopeptide transporter, small intestine is... 70 2e-11
PET1_RAT (P51574) Oligopeptide transporter, small intestine isof... 67 8e-11
OPT3_CAEEL (O01840) Putative oligopeptide transporter opt-3 66 2e-10
PET2_HUMAN (Q16348) Oligopeptide transporter, kidney isoform (Pe... 65 5e-10
PET2_RABIT (P46029) Oligopeptide transporter, kidney isoform (Pe... 64 7e-10
PET2_RAT (Q63424) Oligopeptide transporter, kidney isoform (Pept... 62 3e-09
OPT1_DROME (P91679) Oligopeptide transporter 1 (YIN protein) 60 1e-08
PT2A_ARATH (P46031) Peptide transporter PTR2-A 57 1e-07
PTR2_YEAST (P32901) Peptide transporter PTR2 (Peptide permease P... 52 3e-06
OPT1_CAEEL (Q17758) Oligopeptide transporter 1 (Di-/tri-peptide ... 51 6e-06
OPT2_CAEEL (Q21219) Oligopeptide transporter 2 (Di-/tri-peptide ... 41 0.008
Y326_METJA (Q57772) Hypothetical protein MJ0326 37 0.15
YBHN_ECOLI (P75770) Hypothetical protein ybhN 33 2.2
YBGH_ECOLI (P75742) Hypothetical transporter ybgH 33 2.2
PTR2_CANAL (P46030) Peptide transporter PTR2 32 2.8
YHCA_BACSU (P54585) Hypothetical transport protein yhcA 32 4.8
>PT2B_ARATH (P46032) Peptide transporter PTR2-B (Histidine
transporting protein)
Length = 585
Score = 323 bits (828), Expect = 6e-88
Identities = 165/405 (40%), Positives = 262/405 (63%), Gaps = 8/405 (1%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + P S+ GADQFD+ + +ER +K SF+NW+ F I IG + + ++LV+IQ+N G+ LG
Sbjct: 172 GGIKPCVSSFGADQFDDTDSRERVRKASFFNWFYFSINIGALVSSSLLVWIQENRGWGLG 231
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
+GIPT+ + ++I F GTPLYR + P GSP+TR+ QV VA+ RK + VP D+ L+E
Sbjct: 232 FGIPTVFMGLAIASFFFGTPLYRFQKPGGSPITRISQVVVASFRKSSVKVPEDATLLYET 291
Query: 173 SIEEYTSKGRYRINHSSSLRFLDKAAV------KTGQTS-PWMLCTVTQIEETKQMTKML 225
+ G +I H+ ++LDKAAV K+G S W LCTVTQ+EE K + +M
Sbjct: 292 QDKNSAIAGSRKIEHTDDCQYLDKAAVISEEESKSGDYSNSWRLCTVTQVEELKILIRMF 351
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
P+ + I S + +Q +T+F++QG ++ +G+ F++PPA L F ++I V +YDR
Sbjct: 352 PIWASGIIFSAVYAQMSTMFVQQGRAMNCKIGS-FQLPPAALGTFDTASVIIWVPLYDRF 410
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPL 345
VP+ R++T +G T +QR+GIGL + V+ M A ++E RL +A + L+ +P+
Sbjct: 411 IVPLARKFTGVDKGFTEIQRMGIGLFVSVLCMAAAAIVEIIRLHMANDLGLVESGAPVPI 470
Query: 346 TIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSI 405
++ +PQ+ ++G A+ F I +LEFFYDQ+P++M+SL ++ A T ++GN+LS+ +L++
Sbjct: 471 SVLWQIPQYFILGAAEVFYFIGQLEFFYDQSPDAMRSLCSALALLTNALGNYLSSLILTL 530
Query: 406 VADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIA 450
V T++NG +GWI +NLN +DY++ LA LS VN +F A
Sbjct: 531 VTYFTTRNGQEGWISDNLNSGHLDYFFWLLAGLSLVNMAVYFFSA 575
>CHL1_ARATH (Q05085) Nitrate/chlorate transporter
Length = 590
Score = 285 bits (730), Expect = 1e-76
Identities = 162/416 (38%), Positives = 252/416 (59%), Gaps = 17/416 (4%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFALG 112
G + + S G+DQFDE E KERS+ F+N + F I +G++ A T+LVY+QD+VG G
Sbjct: 161 GGVKASVSGFGSDQFDETEPKERSKMTYFFNRFFFCINVGSLLAVTVLVYVQDDVGRKWG 220
Query: 113 YGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEV 172
YGI +V+++ VF+ GT YR + GSP+T++ V VAA R KL +P D L++V
Sbjct: 221 YGICAFAIVLALSVFLAGTNRYRFKKLIGSPMTQVAAVIVAAWRNRKLELPADPSYLYDV 280
Query: 173 S---IEEYTSKGRYRINHSSSLRFLDKAAVK---TGQTS----PWMLCTVTQIEETKQMT 222
E + KG+ ++ H+ R LDKAA++ G TS W L T+T +EE KQ+
Sbjct: 281 DDIIAAEGSMKGKQKLPHTEQFRSLDKAAIRDQEAGVTSNVFNKWTLSTLTDVEEVKQIV 340
Query: 223 KMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIY 282
+MLP+ T + T+ +Q TTL + Q TLDR +G+ FEIPPA + F +L++ +Y
Sbjct: 341 RMLPIWATCILFWTVHAQLTTLSVAQSETLDRSIGS-FEIPPASMAVFYVGGLLLTTAVY 399
Query: 283 DRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGP-LD 341
DRV + + ++ P G+ LQR+G+GL + M VA L+E KRL A + GP +
Sbjct: 400 DRVAIRLCKKLFNYPHGLRPLQRIGLGLFFGSMAMAVAALVELKRLRTAHAH---GPTVK 456
Query: 342 TIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTF 401
T+PL ++L+PQ+ ++GI + + +L+FF + P+ MK + T +TL++G F S+
Sbjct: 457 TLPLGFYLLIPQYLIVGIGEALIYTGQLDFFLRECPKGMKGMSTGLLLSTLALGFFFSSV 516
Query: 402 LLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYND 457
L++IV T K H WI ++LN R+ +Y +A+L A+NF+ F V +K++VY +
Sbjct: 517 LVTIVEKFTGK-AHP-WIADDLNKGRLYNFYWLVAVLVALNFLIFLVFSKWYVYKE 570
>PTR2_SCHPO (Q9P380) Probable peptide transporter Ptr2 (Peptide
permease Ptr2)
Length = 618
Score = 75.9 bits (185), Expect = 2e-13
Identities = 95/410 (23%), Positives = 164/410 (39%), Gaps = 66/410 (16%)
Query: 84 WWVFY--ILIGTISAQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRLPSG 141
+ +FY I +G++S ++ GF Y +P + V+ +++ + Y+H PSG
Sbjct: 247 YMIFYWSINVGSLSVLAT-TSLESTKGFVYAYLLPLCVFVIPLIILAVSKRFYKHTPPSG 305
Query: 142 SPLTRMVQVF-VAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVK 200
S R+ QVF +AA K+ L S T+ GR + F+D
Sbjct: 306 SIFVRVGQVFFLAAQNKFNLEKTKPSCT---------TTVGRVTLKDQWDDLFID----- 351
Query: 201 TGQTSPWMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHF 260
E K+ + + I Q T I Q + G
Sbjct: 352 ----------------ELKRALRACKTFLFYPIYWVCYGQMTNNLISQAGQMQTG----- 390
Query: 261 EIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVA 320
+ AF +I ++I + I D + P++R+Y + I R+ +G + MI A
Sbjct: 391 NVSNDLFQAFDSIALIIFIPICDNIIYPLLRKYNIPFKPIL---RITLGFMFATASMIYA 447
Query: 321 CLIERKRLSVAR-ENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPES 379
+++ K N + ++++I +P + L+ ++ F I LEF + +AP S
Sbjct: 448 AVLQAKIYQRGPCYANFTDTCVSNDISVWIQIPAYVLIAFSEIFASITGLEFAFTKAPPS 507
Query: 380 MKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLS 439
MKS+ T+ T + G +LSI T+ N W+ + V+ AF+A
Sbjct: 508 MKSIITALFLFTNAFG-----AILSICISSTAVNPKLTWMYTGIAVT------AFIA--G 554
Query: 440 AVNFICFFVIAKF--------FVYNDDVTRTKMDLEMNPNSSKHKAEISQ 481
+ ++CF F ND + TK D+E + S A+ SQ
Sbjct: 555 IMFWVCFHHYDAMEDEQNQLEFKRNDAL--TKKDVEKEVHDSYSMADESQ 602
>PET1_RABIT (P36836) Oligopeptide transporter, small intestine
isoform (Peptide transporter 1) (Intestinal H(+)/peptide
cotransporter)
Length = 707
Score = 69.7 bits (169), Expect = 2e-11
Identities = 68/283 (24%), Positives = 126/283 (44%), Gaps = 51/283 (18%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQ----TILVYIQD--- 105
G + P S G DQF+E + K+R++ S +FY+ I S T +V +Q
Sbjct: 137 GGIKPCVSAFGGDQFEEGQEKQRNRFFS-----IFYLAINAGSLLSTIITPMVRVQQCGI 191
Query: 106 ---NVGFALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNV 162
+ L +GIP IL+ VS++VF++G+ +Y+ P G+ L+++V+ A++
Sbjct: 192 HVKQACYPLAFGIPAILMAVSLIVFIIGSGMYKKFKPQGNILSKVVKCICFAIK------ 245
Query: 163 PIDSKELHEVSIEEYTSKGRYRINH-SSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQM 221
++ R+R +LD A K + I + K +
Sbjct: 246 ----------------NRFRHRSKQFPKRAHWLDWAKEKYDE---------RLIAQIKMV 280
Query: 222 TKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVI 281
T++L + I + + Q + + Q TT+ +G EI P + I ++I V I
Sbjct: 281 TRVLFLYIPLPMFWALFDQQGSRWTLQATTMSGRIGI-LEIQPDQMQTVNTILIIILVPI 339
Query: 282 YDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIE 324
D V P+I + N T L+++ IG+ + + + A +++
Sbjct: 340 MDAVVYPLIAKCGLN---FTSLKKMTIGMFLASMAFVAAAILQ 379
Score = 42.4 bits (98), Expect = 0.003
Identities = 36/128 (28%), Positives = 58/128 (45%), Gaps = 20/128 (15%)
Query: 351 VPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLT 410
+PQ+ L+ + I LEF Y QAP +MKS+ + T+++GN ++ IVA
Sbjct: 584 IPQYFLITSGEVVFSITGLEFSYSQAPSNMKSVLQAGWLLTVAVGN----IIVLIVAGAG 639
Query: 411 SKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYNDDVTRTKMDLEMNP 470
N K W Y F ALL V + F ++A+F+ Y V +++ +
Sbjct: 640 QIN--KQWA----------EYILFAALLLVV-CVIFAIMARFYTY---VNPAEIEAQFEE 683
Query: 471 NSSKHKAE 478
+ K E
Sbjct: 684 DEKKKNPE 691
>PET1_HUMAN (P46059) Oligopeptide transporter, small intestine
isoform (Peptide transporter 1) (Intestinal H(+)/peptide
cotransporter) (Solute carrier family 15, member 1)
Length = 708
Score = 69.7 bits (169), Expect = 2e-11
Identities = 65/284 (22%), Positives = 123/284 (42%), Gaps = 53/284 (18%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVG---- 108
G + P S G DQF+E + K+R++ S +FY+ I S + ++ V
Sbjct: 137 GGIKPCVSAFGGDQFEEGQEKQRNRFFS-----IFYLAINAGSLLSTIITPMLRVQQCGI 191
Query: 109 ------FALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNV 162
+ L +G+P L+ V+++VFVLG+ +Y+ P G+ + ++ + A++
Sbjct: 192 HSKQACYPLAFGVPAALMAVALIVFVLGSGMYKKFKPQGNIMGKVAKCIGFAIKN----- 246
Query: 163 PIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQ--IEETKQ 220
R H S KA K W + I + K
Sbjct: 247 ---------------------RFRHRS------KAFPKREHWLDWAKEKYDERLISQIKM 279
Query: 221 MTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVV 280
+T+++ + I + + Q + + Q TT+ +GA EI P + I ++I V
Sbjct: 280 VTRVMFLYIPLPMFWALFDQQGSRWTLQATTMSGKIGA-LEIQPDQMQTVNAILIVIMVP 338
Query: 281 IYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIE 324
I+D V P+I + N T L+++ +G+V+ + +VA +++
Sbjct: 339 IFDAVLYPLIAKCGFN---FTSLKKMAVGMVLASMAFVVAAIVQ 379
Score = 43.1 bits (100), Expect = 0.002
Identities = 38/159 (23%), Positives = 74/159 (45%), Gaps = 21/159 (13%)
Query: 322 LIERKRLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMK 381
+++RK S E + + + + + +PQ+ L+ + + LEF Y QAP +MK
Sbjct: 557 IVQRKNDSCP-EVKVFEDISANTVNMALQIPQYFLLTCGEVVFSVTGLEFSYSQAPSNMK 615
Query: 382 SLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAV 441
S+ + T+++GN ++ IVA + K W Y F ALL V
Sbjct: 616 SVLQAGWLLTVAVGN----IIVLIVAG--AGQFSKQWA----------EYILFAALLLVV 659
Query: 442 NFICFFVIAKFFVYNDDVTRTKMDLEMNPNSSKHKAEIS 480
+ F ++A+F+ Y + +++ + + + K++ E S
Sbjct: 660 -CVIFAIMARFYTY---INPAEIEAQFDEDEKKNRLEKS 694
>PET1_RAT (P51574) Oligopeptide transporter, small intestine isoform
(Peptide transporter 1) (Intestinal H(+)/peptide
cotransporter) (Proton-coupled dipeptide cotransporter)
Length = 710
Score = 67.4 bits (163), Expect = 8e-11
Identities = 68/282 (24%), Positives = 125/282 (44%), Gaps = 49/282 (17%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISA-QTILVYI-------- 103
G + P S G DQF+E + K+R++ S +FY+ I S TI+ I
Sbjct: 137 GGIKPCVSAFGGDQFEEGQEKQRNRFFS-----IFYLAINAGSLLSTIITPILRVQQCGI 191
Query: 104 -QDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNV 162
+ L +G+P L+ V+++VFVLG+ +Y+ P G+ + ++ + A++
Sbjct: 192 HSQQACYPLAFGVPAALMAVALIVFVLGSGMYKKFQPQGNIMGKVAKCIRFAIK------ 245
Query: 163 PIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMT 222
+ + SK + NH +LD A K + I + K MT
Sbjct: 246 ----------NRFRHRSKAFPKRNH-----WLDWAKEKYDE---------RLISQIKIMT 281
Query: 223 KMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIY 282
K++ + I + + Q + + Q TT+ +G EI P + I ++I V I
Sbjct: 282 KVMFLYIPLPMFWALFDQQGSRWTLQATTMTGKIGT-IEIQPDQMQTVNAILIVIMVPIV 340
Query: 283 DRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIE 324
D V P+I + N T L+++ +G+ + + +VA +++
Sbjct: 341 DAVVYPLIAKCGFN---FTSLKKMTVGMFLASMAFVVAAIVQ 379
Score = 43.5 bits (101), Expect = 0.001
Identities = 31/107 (28%), Positives = 49/107 (44%), Gaps = 21/107 (19%)
Query: 351 VPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLT 410
+PQ+ L+ + + LEF Y QAP +MKS+ + T++IGN I+ +
Sbjct: 587 IPQYFLLTCGEVVFSVTGLEFSYSQAPSNMKSVLQAGWLLTVAIGN--------IIVLIV 638
Query: 411 SKNGH--KGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVY 455
++ GH K W Y A L V I F ++A+F+ Y
Sbjct: 639 AEAGHFDKQWA-----------EYVLFASLLLVVCIIFAIMARFYTY 674
>OPT3_CAEEL (O01840) Putative oligopeptide transporter opt-3
Length = 701
Score = 66.2 bits (160), Expect = 2e-10
Identities = 63/282 (22%), Positives = 124/282 (43%), Gaps = 47/282 (16%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVG---- 108
G + P S ADQF E + RSQ F++++ F I G++ A I ++ V
Sbjct: 128 GCIKPCVSAFAADQFTEDQKDLRSQ---FFSFFYFAINGGSLFAIIITPILRGRVQCFGN 184
Query: 109 ---FALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPL-TRMVQVFVAAMRKWKLNVPI 164
F L +G+P +L+++++++F++G +Y+ PS + +++V V ++RK
Sbjct: 185 AHCFPLAFGVPGVLMLLALILFLMGWSMYKKHPPSKENVGSKVVAVIYTSLRKMVGGASR 244
Query: 165 DSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKM 224
D H +LD AA + Q I+ T+ + +
Sbjct: 245 DKPVTH----------------------WLDHAAPEHSQ---------KMIDSTRGLLNV 273
Query: 225 LPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDR 284
+ + Q + ++ Q LD +G HF I P + A + +LI V I++
Sbjct: 274 AVIFCPLIFFWALFDQQGSTWVLQARRLDGRVG-HFSILPEQIHAINPVCVLILVPIFEG 332
Query: 285 VFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERK 326
P +R+ T+ +T L+++ +G ++ +A +++ K
Sbjct: 333 WVYPALRKITR----VTPLRKMAVGGLLTAFSFAIAGVLQLK 370
Score = 35.4 bits (80), Expect = 0.33
Identities = 16/58 (27%), Positives = 32/58 (54%)
Query: 345 LTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFL 402
++I +PQ+ ++ + + + + LEF Y QA +MKS+ T+ T+ GN + +
Sbjct: 573 VSILWSLPQYIIITLGEVLLSVTGLEFAYSQAAPNMKSVLTAMWLLTVFAGNLIDMMI 630
>PET2_HUMAN (Q16348) Oligopeptide transporter, kidney isoform
(Peptide transporter 2) (Kidney H(+)/peptide
cotransporter) (Solute carrier family 15, member 2)
Length = 729
Score = 64.7 bits (156), Expect = 5e-10
Identities = 69/311 (22%), Positives = 138/311 (44%), Gaps = 54/311 (17%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILI--GTISAQTILVYIQDNVG-- 108
G + P + G DQF+E +ER++ S VFY+ I G++ + I ++ +V
Sbjct: 158 GGIKPCVAAFGGDQFEEKHAEERTRYFS-----VFYLSINAGSLISTFITPMLRGDVQCF 212
Query: 109 ----FALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPI 164
+AL +G+P +L+V++++VF +G+ +Y P G+ + ++ + A+ N
Sbjct: 213 GEDCYALAFGVPGLLMVIALVVFAMGSKIYNKPPPEGNIVAQVFKCIWFAISNRFKNRSG 272
Query: 165 DSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKM 224
D + H+ +LD AA K P L I + K +T++
Sbjct: 273 DIPKRHD---------------------WLDWAAEK----YPKQL-----IMDVKALTRV 302
Query: 225 LPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDR 284
L + I + ++ Q + + Q ++R +G F + P + + +LI + ++D
Sbjct: 303 LFLYIPLPMFWALLDQQGSRWTLQAIRMNRNLG-FFVLQPDQMQVLNPLLVLIFIPLFDF 361
Query: 285 VFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIP 344
V + R +K + L+++ +G+++ + VA +E K N + P P
Sbjct: 362 V---IYRLVSKCGINFSSLRKMAVGMILACLAFAVAARVEIK-------INEMAPAQPGP 411
Query: 345 LTIFILVPQFA 355
+F+ V A
Sbjct: 412 QEVFLQVLNLA 422
Score = 40.4 bits (93), Expect = 0.010
Identities = 18/48 (37%), Positives = 29/48 (59%)
Query: 351 VPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFL 398
+PQ+AL+ + + LEF Y QAP SMKS+ + T+++GN +
Sbjct: 612 LPQYALVTAGEVMFSVTGLEFSYSQAPSSMKSVLQAAWLLTIAVGNII 659
>PET2_RABIT (P46029) Oligopeptide transporter, kidney isoform
(Peptide transporter 2) (Kidney H(+)/peptide
cotransporter)
Length = 729
Score = 64.3 bits (155), Expect = 7e-10
Identities = 61/281 (21%), Positives = 130/281 (45%), Gaps = 45/281 (16%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVG---- 108
G + P + G DQF+E +ER++ +++ + I G++ + I ++ +V
Sbjct: 158 GGIKPCVAAFGGDQFEEKHAEERTR---YFSGFYLAINAGSLISTFITPMLRGDVQCFGE 214
Query: 109 --FALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQ-VFVAAMRKWKLNVPID 165
+AL +G+P +L+V++++VF +G+ +Y+ P G+ + ++V+ ++ A ++K
Sbjct: 215 DCYALAFGVPGLLMVIALVVFAMGSKMYKKPPPEGNIVAQVVKCIWFAISNRFKNRSEDI 274
Query: 166 SKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKML 225
K H +LD AA K P L I + K +T++L
Sbjct: 275 PKRQH----------------------WLDWAAEK----YPKQL-----IMDVKTLTRVL 303
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
+ I + ++ Q + + Q T ++ +G F + P + + +LI + ++D V
Sbjct: 304 FLYIPLPMFWALLDQQGSRWTLQATKMNGNLG-FFVLQPDQMQVLNPLLVLIFIPLFDLV 362
Query: 286 FVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERK 326
+ R +K T L+++ +G+V+ + A +E K
Sbjct: 363 ---IYRLISKCGINFTSLRKMAVGMVLACLAFAAAATVEIK 400
Score = 40.4 bits (93), Expect = 0.010
Identities = 19/48 (39%), Positives = 29/48 (59%)
Query: 351 VPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFL 398
+PQ+AL+ + + LEF Y QAP SMKS+ + T++IGN +
Sbjct: 612 LPQYALVTAGEVMFSVTGLEFSYSQAPSSMKSVLQAAWLLTVAIGNII 659
>PET2_RAT (Q63424) Oligopeptide transporter, kidney isoform (Peptide
transporter 2) (Kidney H(+)/peptide cotransporter)
Length = 729
Score = 62.0 bits (149), Expect = 3e-09
Identities = 63/286 (22%), Positives = 129/286 (45%), Gaps = 55/286 (19%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILI--GTISAQTILVYIQDNVG-- 108
G + P + G DQF+E + R++ S VFY+ I G++ + I ++ +V
Sbjct: 158 GGIKPCVAAFGGDQFEEEHAEARTRYFS-----VFYLAINAGSLISTFITPMLRGDVKCF 212
Query: 109 ----FALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPI 164
+AL +G+P +L+V++++VF +G+ +YR P G+ + ++++ W
Sbjct: 213 GQDCYALAFGVPGLLMVLALVVFAMGSKMYRKPPPEGNIVAQVIKCI------W------ 260
Query: 165 DSKELHEVSIEEYTSKGRYRINHSSSL----RFLDKAAVKTGQTSPWMLCTVTQIEETKQ 220
+ R+R N S L +LD AA K + I + K
Sbjct: 261 ------------FALCNRFR-NRSGDLPKRQHWLDWAAEKYPK---------HLIADVKA 298
Query: 221 MTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVV 280
+T++L + I + ++ Q + + Q ++ +G F + P + +LI +
Sbjct: 299 LTRVLFLYIPLPMFWALLDQQGSRWTLQANKMNGDLG-FFVLQPDQMQVLNPFLVLIFIP 357
Query: 281 IYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERK 326
++D V +I + N + L+++ +G+++ + VA L+E K
Sbjct: 358 LFDLVIYRLISKCRIN---FSSLRKMAVGMILACLAFAVAALVETK 400
Score = 40.8 bits (94), Expect = 0.008
Identities = 20/54 (37%), Positives = 32/54 (59%)
Query: 345 LTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFL 398
L+I +PQ+ L+ A+ + LEF Y QAP SMKS+ + T+++GN +
Sbjct: 606 LSIAWQLPQYVLVTAAEVMFSVTGLEFSYSQAPSSMKSVLQAAWLLTVAVGNII 659
>OPT1_DROME (P91679) Oligopeptide transporter 1 (YIN protein)
Length = 737
Score = 60.5 bits (145), Expect = 1e-08
Identities = 66/308 (21%), Positives = 130/308 (41%), Gaps = 49/308 (15%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVG---- 108
G ++P S G +QF + Q F++ + F I G++ + T ++ +V
Sbjct: 140 GGINPCVSPFGGEQFSL--PAQSFQLAKFFSLFYFAINAGSLISTTFTPILRADVHCFGD 197
Query: 109 ---FALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPID 165
F+L +G+P IL++ S+++F+ G LYR + P+G+ + + + A + W+
Sbjct: 198 QDCFSLAFGVPAILMIFSVIIFMAGKRLYRCQPPAGNMIFGVSRCIADAFKGWQ------ 251
Query: 166 SKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKML 225
K H +E FLD A G ++ETK + ++L
Sbjct: 252 -KRRHSEPMES----------------FLDYAKPTVGS---------RMVQETKCLGRIL 285
Query: 226 PVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDRV 285
+ + + + Q + + Q T +D G F+I P + + +L + ++D +
Sbjct: 286 RLFLPFPVFWALFDQQGSRWTFQATRMD-GNVLGFQIKPDQMQVVNPLLILGFLPLFDYI 344
Query: 286 FVPVIRRY-TKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIP 344
P + R + P LQ+L +G ++ + ++ +E K A + P D
Sbjct: 345 IYPALARCGIRRP-----LQKLTLGFLLAALGFFLSAGLEMKMEQAAYRATPIEP-DMTH 398
Query: 345 LTIFILVP 352
L I+ +P
Sbjct: 399 LRIYNGMP 406
Score = 42.7 bits (99), Expect = 0.002
Identities = 31/116 (26%), Positives = 53/116 (44%), Gaps = 19/116 (16%)
Query: 344 PLTIFIL--VPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTF 401
P T+ IL +PQ +M A+ + LEF Y Q+P SMKS+ + +++IGN
Sbjct: 582 PSTVSILWQLPQIVVMTAAEVMFSVTGLEFSYSQSPPSMKSVLQACWLLSVAIGN----M 637
Query: 402 LLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYND 457
L+ ++A+ + G + A L V+ + F +A+ + Y D
Sbjct: 638 LVVVIAEFKFTSSQSG-------------EFTLFASLMLVDMMIFLWLARSYQYKD 680
>PT2A_ARATH (P46031) Peptide transporter PTR2-A
Length = 610
Score = 57.0 bits (136), Expect = 1e-07
Identities = 57/222 (25%), Positives = 100/222 (44%), Gaps = 27/222 (12%)
Query: 269 AFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRL 328
AF +I ++I + I D + P++R+Y + I R+ +G + MI A +++ K
Sbjct: 399 AFDSIALIIFIPICDNIIYPLLRKYNIPFKPIL---RITLGFMFATASMIYAAVLQAKIY 455
Query: 329 SVAR-ENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSY 387
N + ++++I +P + L+ ++ F I LEF + +AP SMKS+ T+
Sbjct: 456 QRGPCYANFTDTCVSNDISVWIQIPAYVLIAFSEIFASITGLEFAFTKAPPSMKSIITAL 515
Query: 388 ATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFF 447
T + G +LSI T+ N W+ + V+ AF+A + ++CF
Sbjct: 516 FLFTNAFG-----AILSICISSTAVNPKLTWMYTGIAVT------AFIA--GIMFWVCFH 562
Query: 448 VIAKF--------FVYNDDVTRTKMDLEMNPNSSKHKAEISQ 481
F ND + TK D+E + S A+ SQ
Sbjct: 563 HYDAMEDEQNQLEFKRNDAL--TKKDVEKEVHDSYSMADESQ 602
>PTR2_YEAST (P32901) Peptide transporter PTR2 (Peptide permease
PTR2)
Length = 601
Score = 52.4 bits (124), Expect = 3e-06
Identities = 71/330 (21%), Positives = 128/330 (38%), Gaps = 49/330 (14%)
Query: 82 YNWWVFYILIGTIS--AQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRLP 139
+ ++ F I +G++S A T L Y + GF Y +P ++++ + G Y R
Sbjct: 244 FMFFYFMINVGSLSLMATTELEY---HKGFWAAYLLPFCFFWIAVVTLIFGKKQYIQRPI 300
Query: 140 SGSPLTRMVQVFVAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAV 199
+ + +V W L +K ++ N + K +V
Sbjct: 301 GDKVIAKSFKVC------WIL------------------TKNKFDFNAA-------KPSV 329
Query: 200 KTGQTSPWMLCTVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAH 259
+ PW ++E K+ V I I T + FI Q + ++
Sbjct: 330 HPEKNYPW---NDKFVDEIKRALAACKVFIFYPIYWTQYGTMISSFITQASMMEL----- 381
Query: 260 FEIPPACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIV 319
IP L AF +I ++I + I+++ P IRRYT + + ++ G + M
Sbjct: 382 HGIPNDFLQAFDSIALIIFIPIFEKFVYPFIRRYTP----LKPITKIFFGFMFGSFAMTW 437
Query: 320 ACLIERKRLSVAR-ENNLLGPLDTIPLTIFILVPQFALMGIADTFVDIAKLEFFYDQAPE 378
A +++ N LG + + +P + L+ ++ F I LE+ Y +AP
Sbjct: 438 AAVLQSFVYKAGPWYNEPLGHNTPNHVHVCWQIPAYVLISFSEIFASITGLEYAYSKAPA 497
Query: 379 SMKSLGTSYATTTLSIGNFLSTFLLSIVAD 408
SMKS S T + G+ + L + D
Sbjct: 498 SMKSFIMSIFLLTNAFGSAIGCALSPVTVD 527
>OPT1_CAEEL (Q17758) Oligopeptide transporter 1 (Di-/tri-peptide
transporter CPTA)
Length = 785
Score = 51.2 bits (121), Expect = 6e-06
Identities = 59/280 (21%), Positives = 118/280 (42%), Gaps = 33/280 (11%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTILVYIQDNVGFA-- 110
G + P S G DQF + S F++ + F I G++ + + Y + F
Sbjct: 145 GGIKPCVSAFGGDQFPAHYTRMISL---FFSMFYFSINAGSLISMWLTPYFRSMSCFGHD 201
Query: 111 ----LGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQVFVAAMRKWKLNVPIDS 166
L +GIP IL++V+ LVF+ G+ Y+ P + + +++ A+RK S
Sbjct: 202 SCYPLAFGIPAILMIVATLVFMAGSFWYKKVPPKENIIFKVIGTITTALRK-----KASS 256
Query: 167 KELHEVS-IEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQ-IEETKQMTKM 224
H+ S EY+ G A+ T + C + IE+ K++ ++
Sbjct: 257 SSTHQRSHWLEYSLDGH-------------DCALSTECKNLHGNCAQRRYIEDIKRLFRV 303
Query: 225 LPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVIYDR 284
+ ++I + + Q + ++ Q +D + FEI P + +L + I+
Sbjct: 304 IVMMIPVPMFWALYDQQGSTWVLQAVGMDAKVFG-FEILPDQMGVLNAFLILFFIPIFQS 362
Query: 285 VFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIE 324
+ P I + +TML+++ G ++ + V +++
Sbjct: 363 IVYPTIEKLGFQ---MTMLRKMAGGGILTAVSFFVCGIVQ 399
Score = 33.5 bits (75), Expect = 1.3
Identities = 18/67 (26%), Positives = 36/67 (52%), Gaps = 2/67 (2%)
Query: 345 LTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLST--FL 402
++I +PQ+ ++ + I LEF Y +A +KS+ + T +IG+ + F+
Sbjct: 668 VSILWQIPQYVILTAGEVLFSITGLEFAYTEASPQLKSVVQALWLFTTAIGDLIVVVIFM 727
Query: 403 LSIVADL 409
L+I +D+
Sbjct: 728 LNIFSDV 734
>OPT2_CAEEL (Q21219) Oligopeptide transporter 2 (Di-/tri-peptide
transporter CPTB)
Length = 835
Score = 40.8 bits (94), Expect = 0.008
Identities = 56/283 (19%), Positives = 125/283 (43%), Gaps = 34/283 (12%)
Query: 53 GELSPTFSTMGADQFDEFEXKERSQKLSFYNWWVFYILIGTISAQTI--LVYIQDNVG-- 108
G + P S G DQF+ +ER L F++ + F I G++ + I + Q +G
Sbjct: 159 GGIKPCVSAFGGDQFEL--GQERMLSL-FFSMFYFSINAGSMISTFISPIFRSQPCLGQD 215
Query: 109 --FALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQ-VFVAAMRKWKL-NVPI 164
+ + +GIP IL++V+ LVF+ G+ Y+ P + + + +F A K K + P
Sbjct: 216 SCYPMAFGIPAILMIVATLVFMGGSFWYKKNPPKDNVFGEVSRLMFRAVGNKMKSGSTPK 275
Query: 165 DSKELHEVSIEEYTSKGRYRINHSSSL--RFLDKAAVKTGQTSPWMLCTVTQ-IEETKQM 221
+ LH Y H +L + L+ A K + LC + I++ + +
Sbjct: 276 EHWLLH------------YLTTHDCALDAKCLELQAEKRNKN----LCQKKKFIDDVRSL 319
Query: 222 TKMLPVLITTCIPSTIISQTTTLFIRQGTTLDRGMGAHFEIPPACLIAFVNIFMLISVVI 281
++L + + + + Q ++++ Q +D + + P + + +L+ + +
Sbjct: 320 LRVLVMFLPVPMFWALYDQQGSVWLIQAIQMDCRLSDTLLLLPDQMQTLNAVLILLFIPL 379
Query: 282 YDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIE 324
+ + PV + + +T L+++ G ++ + ++ ++
Sbjct: 380 FQVIIYPVAAKCVR----LTPLRKMVTGGLLASLAFLITGFVQ 418
Score = 30.8 bits (68), Expect = 8.2
Identities = 26/112 (23%), Positives = 47/112 (41%), Gaps = 16/112 (14%)
Query: 345 LTIFILVPQFALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLS 404
++I +PQ ++ A+ I EF Y Q+ SMK+L + T + G+ S
Sbjct: 695 VSILWQIPQIVVITAAEILFSITGYEFAYSQSAPSMKALVQALWLLTTAAGD-------S 747
Query: 405 IVADLTSKNGHKGWILNNLNVSRIDYYYAFLALLSAVNFICFFVIAKFFVYN 456
I+ +T N + N+ V Y A +++ + F ++ YN
Sbjct: 748 IIVVITILN-----LFENMAVEFFVYAAAMFVVIAIFALLSIF----YYTYN 790
>Y326_METJA (Q57772) Hypothetical protein MJ0326
Length = 436
Score = 36.6 bits (83), Expect = 0.15
Identities = 71/254 (27%), Positives = 108/254 (41%), Gaps = 31/254 (12%)
Query: 174 IEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLCTVTQIEETKQMTKMLPVLITTCI 233
+E+Y +Y N L+ A + T T +++ QI T M V++ TCI
Sbjct: 6 VEKYFEFEKYGTN----LKVETLAGITTFMTMAYIIFVNPQILSTAGMD-FGAVMVATCI 60
Query: 234 PSTIISQTTTLFIRQGTTLDRGMG--AHFEIPPACL-------IAFVNIFMLISVVIYDR 284
S I + L+ R L GMG A+F CL +A +F IS V++
Sbjct: 61 ASAIATLVMGLYARYPFALAPGMGLNAYFTY-GVCLGMGIDWRVALGAVF--ISGVLFII 117
Query: 285 VFVPVIRRYTKN--PRGITMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDT 342
+ + IR + N P I +GIGL + I + A +I + ++ NL+ P
Sbjct: 118 LTLTKIRTWIFNVIPNAIKYGTAVGIGLFIAFIGLKSAGIIVSSKATLVTLGNLMEPSTL 177
Query: 343 IPL-TIF---ILVPQ----FALMGIADTFVDIAKLEFFYDQAPESMKSLGTSYATT--TL 392
+ L IF ILV + L+GI T + + PE + S+ S A T L
Sbjct: 178 LALFGIFLTSILVSRNVIGAILIGIIVT--SLIGMILGISPFPEGIFSMPPSIAPTFLQL 235
Query: 393 SIGNFLSTFLLSIV 406
I L+ LL+IV
Sbjct: 236 DIMGALNLGLLTIV 249
>YBHN_ECOLI (P75770) Hypothetical protein ybhN
Length = 318
Score = 32.7 bits (73), Expect = 2.2
Identities = 37/150 (24%), Positives = 64/150 (42%), Gaps = 9/150 (6%)
Query: 301 TMLQRLGIGLVMHVIVMIVACLIERKRLSVARENNLLGPLDTIPLT-IFILVPQFALMGI 359
T L+ LGIGL+M + V + C + R + L+ P L + I + +MG
Sbjct: 162 TTLRILGIGLLMIIAVYLWFCAFAKHRHMTIKGQKLVLPSWKFALAQMLISSVNWMVMGA 221
Query: 360 ADTFVDIAKLEFFYDQAPESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWI 419
+ + +F+ + S+ IG + F+ + + TS KG I
Sbjct: 222 IIWLLLGQSVNYFFVLGVLLVSSIAGVIVHIPAGIGVLEAVFIALLAGEHTS----KGTI 277
Query: 420 LNNLNVSRIDYYYAFLALLSAVNFICFFVI 449
+ L R+ YY F+ LL A+ IC+ ++
Sbjct: 278 IAALLAYRVLYY--FIPLLLAL--ICYLLL 303
>YBGH_ECOLI (P75742) Hypothetical transporter ybgH
Length = 493
Score = 32.7 bits (73), Expect = 2.2
Identities = 63/330 (19%), Positives = 130/330 (39%), Gaps = 47/330 (14%)
Query: 91 IGTISAQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPLTRMVQV 150
+G+I A Y Q+ +A+G+G+ + ++ +++F+ G + H + R
Sbjct: 149 VGSIIAPIACGYAQEEYSWAMGFGLAAVGMIAGLVIFLCGNRHFTHTRGVNKKVLRATN- 207
Query: 151 FVAAMRKWKLNVPIDSKELHEVSIEEYTSKGRYRINHSSSLRFLDKAAVKTGQTSPWMLC 210
F+ W L + + + L ++I + Y A + +L
Sbjct: 208 FLLPNWGWLLVLLVATPAL--ITILFWKEWSVY-------------ALIVATIIGLGVLA 252
Query: 211 TVTQIEETKQMTKMLPVLITTCIPSTIISQTTTLFIRQGTT-----LDRGMGAHF--EIP 263
+ + E ++ K L +++T T S F +QG + +DR +
Sbjct: 253 KIYRKAENQKQRKELGLIVTL----TFFSMLFWAFAQQGGSSISLYIDRFVNRDMFGYTV 308
Query: 264 PACLIAFVNIFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLI 323
P + +N F +V++ V++ R + + + +GL + + L
Sbjct: 309 PTAMFQSINAF---AVMLCGVFLAWVVKESVAGNRTVRIWGKFALGLGLMSAGFCILTL- 364
Query: 324 ERKRLSVARENNLLGPLDTIPLTIFILVPQFALMGIADTFVD-IAKLEFFYDQAPESMKS 382
AR + + G ++PL + L A+MG A+ F+D +A + + P
Sbjct: 365 ------SARWSAMYGH-SSLPLMVLGL----AVMGFAELFIDPVAMSQITRIEIPGVTGV 413
Query: 383 LGTSYATTTLSIGNFLSTFLLSIVADLTSK 412
L Y + +I N+L+ ++AD TS+
Sbjct: 414 LTGIYMLLSGAIANYLA----GVIADQTSQ 439
>PTR2_CANAL (P46030) Peptide transporter PTR2
Length = 623
Score = 32.3 bits (72), Expect = 2.8
Identities = 18/66 (27%), Positives = 33/66 (49%), Gaps = 1/66 (1%)
Query: 86 VFYILIGTIS-AQTILVYIQDNVGFALGYGIPTILLVVSILVFVLGTPLYRHRLPSGSPL 144
VFY+ I + Q Y + VGF L + +P IL ++ + + P + + P G +
Sbjct: 253 VFYLAINIGAFLQIATSYCERRVGFWLAFFVPMILYIIVPIFLFIVKPKLKIKPPQGQVM 312
Query: 145 TRMVQV 150
T +V++
Sbjct: 313 TNVVKI 318
>YHCA_BACSU (P54585) Hypothetical transport protein yhcA
Length = 532
Score = 31.6 bits (70), Expect = 4.8
Identities = 47/210 (22%), Positives = 86/210 (40%), Gaps = 23/210 (10%)
Query: 273 IFMLISVVIYDRVFVPVIRRYTKNPRGITMLQRLGIGLVMHVIVMIVACLIERKRLSV-- 330
I ++I+V +Y +F+ I Y +N G T LQ G+ L+ IVM++ I
Sbjct: 274 INIIITVALYTGMFLLPI--YLQNLVGFTALQS-GLLLLPGAIVMLIMSPISGILFDKFG 330
Query: 331 ARENNLLGPLDTI-------------PLTIFILVPQFALMGIADTFVDIAKLEFFYDQAP 377
R ++G L T+ P T +L+ G++ + + +Q P
Sbjct: 331 PRPLAIIGLLVTVVTTYQYTQLTIDTPYTHIMLIYSIRAFGMSLLMMPVMTAGM--NQLP 388
Query: 378 ESMKSLGTSYATTTLSIGNFLSTFLLSIVADLTSKNGHKGWILNNLNVSRIDYYYAFLAL 437
+ S GT+ + T I + T L++ + H I + + + ++ +AF
Sbjct: 389 ARLNSHGTAMSNTLRQISGSIGTSLITTIY-TNRTTFHYSQIADKTSTADPNFLHAFQNA 447
Query: 438 LS--AVNFICFFVIAKFFVYNDDVTRTKMD 465
+S VN + AK +VY+ +D
Sbjct: 448 VSNLMVNMNVSYDTAKTYVYSHIYKHASLD 477
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.140 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,896,979
Number of Sequences: 164201
Number of extensions: 1911901
Number of successful extensions: 6385
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 6335
Number of HSP's gapped (non-prelim): 37
length of query: 485
length of database: 59,974,054
effective HSP length: 114
effective length of query: 371
effective length of database: 41,255,140
effective search space: 15305656940
effective search space used: 15305656940
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC147517.7


