

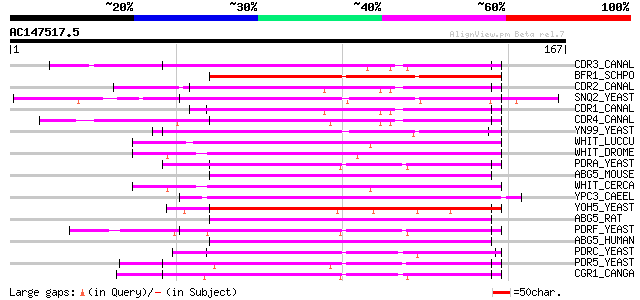
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147517.5 + phase: 0
(167 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CDR3_CANAL (O42690) Opaque-specific ABC transporter CDR3 70 3e-12
BFR1_SCHPO (P41820) Brefeldin A resistance protein 66 3e-11
CDR2_CANAL (P78595) Multidrug resistance protein CDR2 64 2e-10
SNQ2_YEAST (P32568) SNQ2 protein 64 2e-10
CDR1_CANAL (P43071) Multidrug resistance protein CDR1 64 2e-10
CDR4_CANAL (O74676) ABC transporter CDR4 63 3e-10
YN99_YEAST (P53756) Probable ATP-dependent transporter YNR070W 62 5e-10
WHIT_LUCCU (Q05360) White protein 62 5e-10
WHIT_DROME (P10090) White protein 62 6e-10
PDRA_YEAST (P51533) ATP-dependent permease PDR10 62 8e-10
ABG5_MOUSE (Q99PE8) ATP-binding cassette, sub-family G, member 5... 62 8e-10
WHIT_CERCA (Q17320) White protein 61 1e-09
YPC3_CAEEL (Q11180) Putative ABC transporter C05D10.3 in chromos... 61 1e-09
YOH5_YEAST (Q08234) Probable ATP-dependent transporter YOL074C/Y... 61 1e-09
ABG5_RAT (Q99PE7) ATP-binding cassette, sub-family G, member 5 (... 60 2e-09
PDRF_YEAST (Q04182) ATP-dependent permease PDR15 60 2e-09
ABG5_HUMAN (Q9H222) ATP-binding cassette, sub-family G, member 5... 60 2e-09
PDRC_YEAST (Q02785) ATP-dependent permease PDR12 60 3e-09
PDR5_YEAST (P33302) Suppressor of toxicity of sporidesmin 59 5e-09
CGR1_CANGA (O74208) ATP-binding cassette transporter CGR1 (Pleom... 58 9e-09
>CDR3_CANAL (O42690) Opaque-specific ABC transporter CDR3
Length = 1501
Score = 69.7 bits (169), Expect = 3e-12
Identities = 42/103 (40%), Positives = 60/103 (57%), Gaps = 3/103 (2%)
Query: 47 NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRV 106
N + + + + IL ++ G +KP +T L+G +GKTTLL AL+ +L + SG
Sbjct: 844 NLTYTVKIKSEERVILNNIDGWVKPGEVTALMGASGAGKTTLLNALSERLTTGVITSGTR 903
Query: 107 TYNGHEM-SEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
NG E+ S F QR+ YV Q DLH+ TVRE L FSAR++
Sbjct: 904 MVNGGELDSSF--QRSIGYVQQQDLHLETSTVREALKFSARLR 944
Score = 47.4 bits (111), Expect = 2e-05
Identities = 37/136 (27%), Positives = 66/136 (48%), Gaps = 5/136 (3%)
Query: 13 VRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQRLNILKDVSGIIKPS 72
V +++L + +A + S T +N ++ ++ N + +ILK + G+IKP
Sbjct: 114 VAYKNLRVYGDA-IESDYQTTVSNGVLKYARNIFNKFRK-DNDDYSFDILKPMEGLIKPG 171
Query: 73 RMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRV-TYNGHEMSEFVP--QRTAAYVDQND 129
+T++LG P +G +T L +A + + G V +Y+G E + Y + +
Sbjct: 172 EVTVVLGRPGAGCSTFLKTIACRTEGFHVADGSVISYDGITQDEIRNHLRGEVVYCAETE 231
Query: 130 LHIGELTVRETLAFSA 145
H LTV ETL F+A
Sbjct: 232 THFPNLTVGETLEFAA 247
>BFR1_SCHPO (P41820) Brefeldin A resistance protein
Length = 1530
Score = 66.2 bits (160), Expect = 3e-11
Identities = 37/88 (42%), Positives = 53/88 (60%), Gaps = 2/88 (2%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
+L V G + P ++T L+G +GKTTLL LA ++D + +G + NG + +R
Sbjct: 900 LLNGVQGFVVPGKLTALMGESGAGKTTLLNVLAQRVDTGV-VTGDMLVNGRGLDSTFQRR 958
Query: 121 TAAYVDQNDLHIGELTVRETLAFSARVQ 148
T YV Q D+HIGE TVRE L FSA ++
Sbjct: 959 TG-YVQQQDVHIGESTVREALRFSAALR 985
Score = 41.2 bits (95), Expect = 0.001
Identities = 26/89 (29%), Positives = 41/89 (45%), Gaps = 4/89 (4%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNG---HEMSEFV 117
IL + + ++LG P SG +T L ++ + G Y+G +M +F
Sbjct: 176 ILSHCHALANAGELVMVLGQPGSGCSTFLRSVTSDTVHYKRVEGTTHYDGIDKADMKKFF 235
Query: 118 PQRTAAYVDQNDLHIGELTVRETLAFSAR 146
P Y +ND+H LT ETL F+A+
Sbjct: 236 PG-DLLYSGENDVHFPSLTTAETLDFAAK 263
>CDR2_CANAL (P78595) Multidrug resistance protein CDR2
Length = 1499
Score = 63.9 bits (154), Expect = 2e-10
Identities = 38/92 (41%), Positives = 53/92 (57%), Gaps = 3/92 (3%)
Query: 55 RKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEM- 113
+K+ IL V G +KP ++T L+G +GKTTLL L+ ++ + G NGH +
Sbjct: 869 KKEDRVILDHVDGWVKPGQITALMGASGAGKTTLLNCLSERVTTGIITDGERLVNGHALD 928
Query: 114 SEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
S F QR+ YV Q D+H+ TVRE L FSA
Sbjct: 929 SSF--QRSIGYVQQQDVHLETTTVREALQFSA 958
Score = 53.1 bits (126), Expect = 3e-07
Identities = 35/121 (28%), Positives = 62/121 (50%), Gaps = 6/121 (4%)
Query: 32 PTFTNFMVNIVESLLNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLA 91
PT TN + +N L P + +ILK + I++P +T++LG P +G +TLL
Sbjct: 139 PTVTNALWKFTTEAINKLKK-PDDSKYFDILKSMDAIMRPGELTVVLGRPGAGCSTLLKT 197
Query: 92 LA-GKLDPKLKFSGRVTYNG---HEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARV 147
+A + ++TY+G H++ + Y + D+H L+V +TL F+AR+
Sbjct: 198 IAVNTYGFHIGKESQITYDGLSPHDIERHY-RGDVIYSAETDVHFPHLSVGDTLEFAARL 256
Query: 148 Q 148
+
Sbjct: 257 R 257
>SNQ2_YEAST (P32568) SNQ2 protein
Length = 1501
Score = 63.5 bits (153), Expect = 2e-10
Identities = 40/97 (41%), Positives = 56/97 (57%), Gaps = 3/97 (3%)
Query: 52 LPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGH 111
+P + +L +VSG P MT L+G +GKTTLL LA + + +G + NG
Sbjct: 862 IPYEGGKRMLLDNVSGYCIPGTMTALMGESGAGKTTLLNTLAQRNVGII--TGDMLVNGR 919
Query: 112 EMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+ +RT YV Q D+HI ELTVRE+L FSAR++
Sbjct: 920 PIDASFERRTG-YVQQQDIHIAELTVRESLQFSARMR 955
Score = 46.2 bits (108), Expect = 3e-05
Identities = 50/178 (28%), Positives = 82/178 (45%), Gaps = 20/178 (11%)
Query: 2 DRVGIDLPTIEVRFEHLN---IEAEAHVGSISLPTFTNFMVNIVESLLNSLHVLPSRKQR 58
D GI + V E ++ ++A A G+ TF N + + ++ + +K R
Sbjct: 119 DEQGIHIRKAGVTIEDVSAKGVDASALEGA----TFGNILC-LPLTIFKGIKAKRHQKMR 173
Query: 59 LNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKL-KFSGRVTYNGHEMSEFV 117
I+ +V+ + + M L+LG P +G ++ L AG++D SG V Y+G E +
Sbjct: 174 -QIISNVNALAEAGEMILVLGRPGAGCSSFLKVTAGEIDQFAGGVSGEVAYDGIPQEEMM 232
Query: 118 PQRTA--AYVDQNDLHIGELTVRETLAF-------SARVQGVG-PQYGWSTRFLYTYI 165
+ A Y + D+H LTV++TL F + RV V +Y S R LY I
Sbjct: 233 KRYKADVIYNGELDVHFPYLTVKQTLDFAIACKTPALRVNNVSKKEYIASRRDLYATI 290
>CDR1_CANAL (P43071) Multidrug resistance protein CDR1
Length = 1501
Score = 63.5 bits (153), Expect = 2e-10
Identities = 38/92 (41%), Positives = 53/92 (57%), Gaps = 3/92 (3%)
Query: 55 RKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEM- 113
+K+ IL V G +KP ++T L+G +GKTTLL L+ ++ + G NGH +
Sbjct: 871 KKEDRVILDHVDGWVKPGQITALMGASGAGKTTLLNCLSERVTTGIITDGERLVNGHALD 930
Query: 114 SEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
S F QR+ YV Q D+H+ TVRE L FSA
Sbjct: 931 SSF--QRSIGYVQQQDVHLPTSTVREALQFSA 960
Score = 46.2 bits (108), Expect = 3e-05
Identities = 28/93 (30%), Positives = 52/93 (55%), Gaps = 5/93 (5%)
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALA-GKLDPKLKFSGRVTYNG---HEMSE 115
+ILK + I++P +T++LG P +G +TLL +A + ++TY+G H++
Sbjct: 168 DILKSMDAIMRPGELTVVLGRPGAGCSTLLKTIAVNTYGFHIGKESQITYDGLSPHDIER 227
Query: 116 FVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+ Y + D+H L+V +TL F+AR++
Sbjct: 228 HY-RGDVIYSAETDVHFPHLSVGDTLEFAARLR 259
>CDR4_CANAL (O74676) ABC transporter CDR4
Length = 1490
Score = 63.2 bits (152), Expect = 3e-10
Identities = 39/86 (45%), Positives = 51/86 (58%), Gaps = 3/86 (3%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEM-SEFVPQ 119
IL VSG +KP ++T L+G +GKTTLL AL+ +L + G NG + S F Q
Sbjct: 864 ILDHVSGWVKPGQVTALMGASGAGKTTLLNALSDRLTTGVVTEGIRLVNGRPLDSSF--Q 921
Query: 120 RTAAYVDQNDLHIGELTVRETLAFSA 145
R+ YV Q DLH+ TVRE L F+A
Sbjct: 922 RSIGYVQQQDLHLETSTVREALEFAA 947
Score = 51.2 bits (121), Expect = 1e-06
Identities = 39/146 (26%), Positives = 73/146 (49%), Gaps = 13/146 (8%)
Query: 10 TIEVRFEHLNIEAEAHVGSISLPTFTNFMVNIVESLLNSL---HVLPSRKQRLNILKDVS 66
TI V +++L GS S + + +VN++ L+ ++L +ILK +
Sbjct: 123 TIGVAYKNLRA-----YGSASDADYQSTLVNLIPKYLSLFFREYILRHTGPTFDILKPMD 177
Query: 67 GIIKPSRMTLLLGPPSSGKTTLLLALAGK-LDPKLKFSGRVTYNG---HEMSEFVPQRTA 122
G+IKP +T++LG P +G +T L +A + + + YN HE+ + +
Sbjct: 178 GLIKPGELTVVLGRPGAGCSTFLKTIASQTYGYHIDKDSVIRYNSLTPHEIKKHY-RGEV 236
Query: 123 AYVDQNDLHIGELTVRETLAFSARVQ 148
Y + + H +LTV +TL F+A+++
Sbjct: 237 VYCAETENHFPQLTVGDTLEFAAKMR 262
>YN99_YEAST (P53756) Probable ATP-dependent transporter YNR070W
Length = 1333
Score = 62.4 bits (150), Expect = 5e-10
Identities = 39/102 (38%), Positives = 56/102 (54%), Gaps = 3/102 (2%)
Query: 47 NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRV 106
N +P + +L VSG P +T L+G +GKTTLL LA + + +G +
Sbjct: 733 NVSFTIPHSSGQRKLLDSVSGYCVPGTLTALIGESGAGKTTLLNTLAQRNVGTI--TGDM 790
Query: 107 TYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+G M +RT YV Q DLH+ ELTV+E+L FSAR++
Sbjct: 791 LVDGLPMDASFKRRTG-YVQQQDLHVAELTVKESLQFSARMR 831
Score = 50.8 bits (120), Expect = 1e-06
Identities = 34/109 (31%), Positives = 57/109 (52%), Gaps = 12/109 (11%)
Query: 44 SLLNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFS 103
+++ + +R + ILK+VS + K M L+LG P +G T+ L + AG+ +F+
Sbjct: 28 TIIKGIRERKNRNKMKIILKNVSLLAKSGEMVLVLGRPGAGCTSFLKSAAGETS---QFA 84
Query: 104 GRVTYNGHEMSEFVPQR--------TAAYVDQNDLHIGELTVRETLAFS 144
G VT GH + +PQ+ Y + D+H LTV++TL F+
Sbjct: 85 GGVT-TGHISYDGIPQKEMMQHYKPDVIYNGEQDVHFPHLTVKQTLDFA 132
>WHIT_LUCCU (Q05360) White protein
Length = 677
Score = 62.4 bits (150), Expect = 5e-10
Identities = 41/113 (36%), Positives = 61/113 (53%), Gaps = 4/113 (3%)
Query: 38 MVNIVESLLNSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLD 97
+VN V+ + + +P K R +++K+V G+ P + ++G +GKTTLL ALA +
Sbjct: 80 LVNRVKGVFCNERHIP--KPRKHLIKNVCGVAYPGELLAVMGSSGAGKTTLLNALAFRSA 137
Query: 98 PKLKFSGRVT--YNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
++ S NGH + Q AYV Q+DL IG LT RE L F A V+
Sbjct: 138 RGVQISPSSVRMLNGHPVDAKEMQARCAYVQQDDLFIGSLTAREHLIFQATVR 190
>WHIT_DROME (P10090) White protein
Length = 687
Score = 62.0 bits (149), Expect = 6e-10
Identities = 43/114 (37%), Positives = 59/114 (51%), Gaps = 6/114 (5%)
Query: 38 MVNIVESLL-NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKL 96
+VN L N H+ RK ++LK+V G+ P + ++G +GKTTLL ALA +
Sbjct: 91 LVNRTRGLFCNERHIPAPRK---HLLKNVCGVAYPGELLAVMGSSGAGKTTLLNALAFRS 147
Query: 97 DPKLKFS--GRVTYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
++ S G NG + Q AYV Q+DL IG LT RE L F A V+
Sbjct: 148 PQGIQVSPSGMRLLNGQPVDAKEMQARCAYVQQDDLFIGSLTAREHLIFQAMVR 201
>PDRA_YEAST (P51533) ATP-dependent permease PDR10
Length = 1564
Score = 61.6 bits (148), Expect = 8e-10
Identities = 38/99 (38%), Positives = 54/99 (54%), Gaps = 2/99 (2%)
Query: 47 NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRV 106
N + +P + + IL +V G +KP +T L+G +GKTTLL LA + L +G V
Sbjct: 927 NLCYDIPIKNGKRRILDNVDGWVKPGTLTALIGASGAGKTTLLDCLAERTTMGL-ITGDV 985
Query: 107 TYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
+G + P R+ Y Q DLH+ TVRE+L FSA
Sbjct: 986 FVDGRPRDQSFP-RSIGYCQQQDLHLKTATVRESLRFSA 1023
Score = 46.6 bits (109), Expect = 3e-05
Identities = 31/91 (34%), Positives = 48/91 (52%), Gaps = 3/91 (3%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDP-KLKFSGRVTYNGHEMSEFVP- 118
ILK + G I P + ++LG P +G TTLL +++ K+ +TYNG E
Sbjct: 195 ILKPMDGCINPGELLVVLGRPGAGCTTLLKSISVNTHGFKISPDTIITYNGFSNKEIKNH 254
Query: 119 -QRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+ Y ++D+HI LTV +TL AR++
Sbjct: 255 YRGEVVYNAESDIHIPHLTVFQTLYTVARLK 285
>ABG5_MOUSE (Q99PE8) ATP-binding cassette, sub-family G, member 5
(Sterolin-1)
Length = 652
Score = 61.6 bits (148), Expect = 8e-10
Identities = 37/85 (43%), Positives = 51/85 (59%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
ILKDVS I+ ++ +LG SGKTTLL A++G+L G V NG E+ Q
Sbjct: 69 ILKDVSLYIESGQIMCILGSSGSGKTTLLDAISGRLRRTGTLEGEVFVNGCELRRDQFQD 128
Query: 121 TAAYVDQNDLHIGELTVRETLAFSA 145
+YV Q+D+ + LTVRETL ++A
Sbjct: 129 CFSYVLQSDVFLSSLTVRETLRYTA 153
>WHIT_CERCA (Q17320) White protein
Length = 679
Score = 61.2 bits (147), Expect = 1e-09
Identities = 42/114 (36%), Positives = 60/114 (51%), Gaps = 6/114 (5%)
Query: 38 MVNIVESLL-NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKL 96
+VN V+ + N H+ RK ++LK+ SG+ P + ++G +GKTTLL A A +
Sbjct: 82 LVNRVKGVFCNERHIPAPRK---HLLKNDSGVAYPGELLAVMGSSGAGKTTLLNASAFRS 138
Query: 97 DPKLKFSGRVT--YNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
++ S NGH + Q AYV Q+DL IG LT RE L F A V+
Sbjct: 139 SKGVQISPSTIRMLNGHPVDAKEMQARCAYVQQDDLFIGSLTAREHLIFQAMVR 192
>YPC3_CAEEL (Q11180) Putative ABC transporter C05D10.3 in chromosome
III
Length = 598
Score = 60.8 bits (146), Expect = 1e-09
Identities = 33/103 (32%), Positives = 60/103 (58%), Gaps = 2/103 (1%)
Query: 52 LPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGH 111
+P R+ + IL +VSG+ + ++ +LG +GKTTL+ L + L G + +G
Sbjct: 1 MPKRRVK-EILHNVSGMAESGKLLAILGSSGAGKTTLMNVLTSRNLTNLDVQGSILIDGR 59
Query: 112 EMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQGVGPQY 154
+++ + +A+V Q+D+ +G +T RE L F AR++ +G QY
Sbjct: 60 RANKWKIREMSAFVQQHDMFVGTMTAREHLQFMARLR-MGDQY 101
>YOH5_YEAST (Q08234) Probable ATP-dependent transporter
YOL074C/YOL075C
Length = 1294
Score = 60.8 bits (146), Expect = 1e-09
Identities = 33/93 (35%), Positives = 57/93 (60%), Gaps = 5/93 (5%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLD----PKLKFSGRVTYNGHEMSEF 116
IL+ V+ I KP + ++GP SGK++LL ++G+L K SG + +N ++SE
Sbjct: 709 ILQSVNAIFKPGMINAIMGPSGSGKSSLLNLISGRLKSSVFAKFDTSGSIMFNDIQVSEL 768
Query: 117 VPQRTAAYVDQNDLH-IGELTVRETLAFSARVQ 148
+ + +YV Q+D H + LTV+ETL ++A ++
Sbjct: 769 MFKNVCSYVSQDDDHLLAALTVKETLKYAAALR 801
Score = 44.7 bits (104), Expect = 1e-04
Identities = 37/120 (30%), Positives = 53/120 (43%), Gaps = 22/120 (18%)
Query: 48 SLHV----LPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFS 103
SLHV + + K ++ S + + ++G SGKTTLL LA K+ L +
Sbjct: 27 SLHVRDLSIVASKTNTTLVNTFSMDLPSGSVMAVMGGSGSGKTTLLNVLASKISGGLTHN 86
Query: 104 GRVTY----NGHEMSEFVPQRT--------------AAYVDQNDLHIGELTVRETLAFSA 145
G + Y G E +E P+R AY+ Q D+ LT RETL F+A
Sbjct: 87 GSIRYVLEDTGSEPNETEPKRAHLDGQDHPIQKHVIMAYLPQQDVLSPRLTCRETLKFAA 146
>ABG5_RAT (Q99PE7) ATP-binding cassette, sub-family G, member 5
(Sterolin-1)
Length = 652
Score = 60.5 bits (145), Expect = 2e-09
Identities = 36/85 (42%), Positives = 50/85 (58%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
ILKDVS I+ + +LG SGKTTLL A++G+L G V NG E+ Q
Sbjct: 69 ILKDVSLYIESGQTMCILGSSGSGKTTLLDAISGRLRRTGTLEGEVFVNGCELRRDQFQD 128
Query: 121 TAAYVDQNDLHIGELTVRETLAFSA 145
+Y+ Q+D+ + LTVRETL ++A
Sbjct: 129 CVSYLLQSDVFLSSLTVRETLRYTA 153
>PDRF_YEAST (Q04182) ATP-dependent permease PDR15
Length = 1529
Score = 60.1 bits (144), Expect = 2e-09
Identities = 36/94 (38%), Positives = 53/94 (56%), Gaps = 2/94 (2%)
Query: 52 LPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGH 111
+P + + IL +V G +KP +T L+G +GKTTLL LA ++ + +G + +G
Sbjct: 893 VPIKGGQRRILNNVDGWVKPGTLTALMGASGAGKTTLLDCLAERVTMGV-ITGNIFVDGR 951
Query: 112 EMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
E P R+ Y Q DLH+ TVRE+L FSA
Sbjct: 952 LRDESFP-RSIGYCQQQDLHLKTATVRESLRFSA 984
Score = 51.6 bits (122), Expect = 8e-07
Identities = 41/136 (30%), Positives = 66/136 (48%), Gaps = 9/136 (6%)
Query: 19 NIEAEAHVGSISLPTFTNFMVNIVESLLNS-LHVLPSRKQR--LNILKDVSGIIKPSRMT 75
N+ A +S + + NIV LL L +L K+ ILK + G + P +
Sbjct: 143 NLSASGDSADVS---YQSTFANIVPKLLTKGLRLLKPSKEEDTFQILKPMDGCLNPGELL 199
Query: 76 LLLGPPSSGKTTLLLALAGKLDP-KLKFSGRVTYNGHEMSEFVP--QRTAAYVDQNDLHI 132
++LG P SG TTLL +++ K+ V+YNG S+ + Y ++D+H+
Sbjct: 200 VVLGRPGSGCTTLLKSISSNSHGFKIAKDSIVSYNGLSSSDIRKHYRGEVVYNAESDIHL 259
Query: 133 GELTVRETLAFSARVQ 148
LTV +TL AR++
Sbjct: 260 PHLTVYQTLFTVARMK 275
>ABG5_HUMAN (Q9H222) ATP-binding cassette, sub-family G, member 5
(Sterolin-1)
Length = 651
Score = 60.1 bits (144), Expect = 2e-09
Identities = 36/85 (42%), Positives = 50/85 (58%)
Query: 61 ILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQR 120
ILKDVS ++ ++ +LG SGKTTLL A++G+L F G V NG + Q
Sbjct: 68 ILKDVSLYVESGQIMCILGSSGSGKTTLLDAMSGRLGRAGTFLGEVYVNGRALRREQFQD 127
Query: 121 TAAYVDQNDLHIGELTVRETLAFSA 145
+YV Q+D + LTVRETL ++A
Sbjct: 128 CFSYVLQSDTLLSSLTVRETLHYTA 152
>PDRC_YEAST (Q02785) ATP-dependent permease PDR12
Length = 1511
Score = 59.7 bits (143), Expect = 3e-09
Identities = 35/99 (35%), Positives = 56/99 (56%), Gaps = 2/99 (2%)
Query: 50 HVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYN 109
+ +P +L DV G +KP +MT L+G +GKTTLL LA +++ + +G + N
Sbjct: 849 YTIPYDGATRKLLSDVFGYVKPGKMTALMGESGAGKTTLLNVLAQRINMGV-ITGDMLVN 907
Query: 110 GHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
+ R+ YV Q D H+ EL+VRE+L F+A ++
Sbjct: 908 AKPLPASF-NRSCGYVAQADNHMAELSVRESLRFAAELR 945
Score = 51.6 bits (122), Expect = 8e-07
Identities = 24/89 (26%), Positives = 50/89 (55%), Gaps = 2/89 (2%)
Query: 60 NILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRVTYNGHEMSEFVPQ 119
NI+++ +G+++ M ++G P +G +T L L+G+ + G +Y+G + SE + +
Sbjct: 162 NIIQNCTGVVESGEMLFVVGRPGAGCSTFLKCLSGETSELVDVQGEFSYDGLDQSEMMSK 221
Query: 120 RT--AAYVDQNDLHIGELTVRETLAFSAR 146
Y + D H ++TV+ET+ F+ +
Sbjct: 222 YKGYVIYCPELDFHFPKITVKETIDFALK 250
>PDR5_YEAST (P33302) Suppressor of toxicity of sporidesmin
Length = 1511
Score = 58.9 bits (141), Expect = 5e-09
Identities = 36/99 (36%), Positives = 54/99 (54%), Gaps = 2/99 (2%)
Query: 47 NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRV 106
N + + + + IL +V G +KP +T L+G +GKTTLL LA ++ + +G +
Sbjct: 873 NLCYEVQIKAETRRILNNVDGWVKPGTLTALMGASGAGKTTLLDCLAERVTMGV-ITGDI 931
Query: 107 TYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
NG + P R+ Y Q DLH+ TVRE+L FSA
Sbjct: 932 LVNGIPRDKSFP-RSIGYCQQQDLHLKTATVRESLRFSA 969
Score = 49.3 bits (116), Expect = 4e-06
Identities = 38/125 (30%), Positives = 63/125 (50%), Gaps = 14/125 (11%)
Query: 34 FTNFMVNIVESLLNSLHVLPSRKQRLN---ILKDVSGIIKPSRMTLLLGPPSSGKTTLLL 90
+ + +VNI +L S R + N ILK + G + P + ++LG P SG TTLL
Sbjct: 145 YQSTVVNIPYKILKSGLRKFQRSKETNTFQILKPMDGCLNPGELLVVLGRPGSGCTTLLK 204
Query: 91 ALAGK-------LDPKLKFSGRVTYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAF 143
+++ D K+ +SG Y+G ++ + + Y + D+H+ LTV ETL
Sbjct: 205 SISSNTHGFDLGADTKISYSG---YSGDDIKKHF-RGEVVYNAEADVHLPHLTVFETLVT 260
Query: 144 SARVQ 148
AR++
Sbjct: 261 VARLK 265
>CGR1_CANGA (O74208) ATP-binding cassette transporter CGR1
(Pleomorphic drug resistance homolog)
Length = 1542
Score = 58.2 bits (139), Expect = 9e-09
Identities = 37/99 (37%), Positives = 52/99 (52%), Gaps = 2/99 (2%)
Query: 47 NSLHVLPSRKQRLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDPKLKFSGRV 106
N + +P + + IL +V G +KP +T L+G +GKTTLL LA + + +G V
Sbjct: 889 NLCYDVPIKTEVRRILNNVDGWVKPGTLTALMGASGAGKTTLLDCLAERTTMGV-ITGDV 947
Query: 107 TYNGHEMSEFVPQRTAAYVDQNDLHIGELTVRETLAFSA 145
NG R+ Y Q DLH+ TVRE+L FSA
Sbjct: 948 MVNGRPRDTSF-SRSIGYCQQQDLHLKTATVRESLRFSA 985
Score = 50.8 bits (120), Expect = 1e-06
Identities = 35/120 (29%), Positives = 62/120 (51%), Gaps = 6/120 (5%)
Query: 33 TFTNFMVNIVESLLNSLHVLPSRKQ-RLNILKDVSGIIKPSRMTLLLGPPSSGKTTLLLA 91
TF N V ++ ++ P+R+ ILK + G++KP + ++LG P SG TTLL +
Sbjct: 147 TFLNLPVKLLNAVWRKAR--PARESDTFRILKPMDGLLKPGELLVVLGRPGSGCTTLLKS 204
Query: 92 LAGKLDP-KLKFSGRVTYNGHEMSEFVP--QRTAAYVDQNDLHIGELTVRETLAFSARVQ 148
++ ++ ++YNG +E + Y + D+H+ LTV +TL AR++
Sbjct: 205 ISSTTHGFQISKDSVISYNGLTPNEIKKHYRGEVVYNAEADIHLPHLTVYQTLVTVARLK 264
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,983,191
Number of Sequences: 164201
Number of extensions: 748269
Number of successful extensions: 5155
Number of sequences better than 10.0: 863
Number of HSP's better than 10.0 without gapping: 495
Number of HSP's successfully gapped in prelim test: 368
Number of HSP's that attempted gapping in prelim test: 4450
Number of HSP's gapped (non-prelim): 994
length of query: 167
length of database: 59,974,054
effective HSP length: 102
effective length of query: 65
effective length of database: 43,225,552
effective search space: 2809660880
effective search space used: 2809660880
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC147517.5


