

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147496.8 + phase: 1 /partial
(120 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
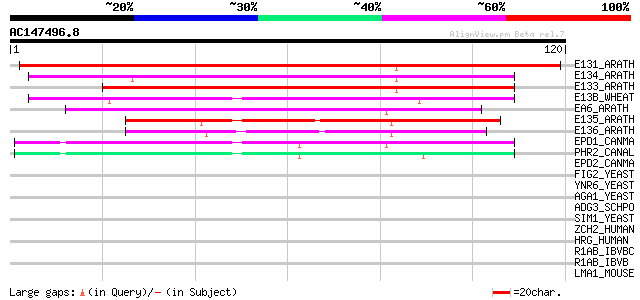
Score E
Sequences producing significant alignments: (bits) Value
E131_ARATH (O65399) Putative glucan endo-1,3-beta-glucosidase 1 ... 99 2e-21
E134_ARATH (Q94CD8) Putative glucan endo-1,3-beta-glucosidase 4 ... 89 2e-18
E133_ARATH (Q9ZU91) Putative glucan endo-1,3-beta-glucosidase 3 ... 87 5e-18
E13B_WHEAT (P52409) Glucan endo-1,3-beta-glucosidase precursor (... 86 1e-17
EA6_ARATH (Q06915) Probable glucan endo-1,3-beta-glucosidase A6 ... 68 4e-12
E135_ARATH (Q9M088) Putative glucan endo-1,3-beta-glucosidase 5 ... 57 1e-08
E136_ARATH (Q93Z08) Putative glucan endo-1,3-beta-glucosidase 6 ... 51 5e-07
EPD1_CANMA (P56092) EPD1 protein precursor (Essential for pseudo... 42 2e-04
PHR2_CANAL (O13318) pH responsive protein 2 precursor (pH-regula... 41 5e-04
EPD2_CANMA (O74137) EPD2 protein precursor (Essential for pseudo... 35 0.030
FIG2_YEAST (P25653) Factor induced gene 2 precursor (Cell wall a... 31 0.43
YNR6_YEAST (P53882) Hypothetical 67.4 kDa protein in RPS3-PSD1 i... 31 0.57
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 30 1.3
ADG3_SCHPO (O74851) Adg3 protein precursor 30 1.3
SIM1_YEAST (P40472) SIM1 protein precursor 29 1.6
ZCH2_HUMAN (Q9C0B9) Zinc finger CCHC domain containing protein 2... 29 2.2
HRG_HUMAN (P04196) Histidine-rich glycoprotein precursor (Histid... 29 2.2
R1AB_IBVBC (Q91QT2) Replicase polyprotein 1ab (pp1ab) (ORF1ab po... 28 2.8
R1AB_IBVB (P27920) Replicase polyprotein 1ab (pp1ab) (ORF1ab pol... 28 2.8
LMA1_MOUSE (P19137) Laminin alpha-1 chain precursor (Laminin A c... 28 2.8
>E131_ARATH (O65399) Putative glucan endo-1,3-beta-glucosidase 1
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta-glucanase)
(Beta-1,3-endoglucanase) (Beta-1,3-glucanase)
Length = 402
Score = 99.0 bits (245), Expect = 2e-21
Identities = 50/118 (42%), Positives = 72/118 (60%), Gaps = 1/118 (0%)
Query: 3 SPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSC 62
+P + ++ T + ++ ++CIA ++LQ ALD+ACG G ++CS IQPG SC
Sbjct: 249 TPVYLLHVSGSGTFLANDTTNQTYCIAMDGVDAKTLQAALDWACGPGRSNCSEIQPGESC 308
Query: 63 YNPNSVHDHASFAFNKYYQK-NPVPNSCNFGGNAVLTNTNPSKASTIYDHGFKLQSDT 119
Y PN+V HASFAFN YYQK SC+F G A++T T+PS S I+ K+ + T
Sbjct: 309 YQPNNVKGHASFAFNSYYQKEGRASGSCDFKGVAMITTTDPSHGSCIFPGSKKVGNRT 366
>E134_ARATH (Q94CD8) Putative glucan endo-1,3-beta-glucosidase 4
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta-glucanase)
(Beta-1,3-endoglucanase) (Beta-1,3-glucanase)
Length = 505
Score = 88.6 bits (218), Expect = 2e-18
Identities = 45/107 (42%), Positives = 62/107 (57%), Gaps = 2/107 (1%)
Query: 5 SSTVPITAPSTSNSPVSSGAS-WCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCY 63
+S P++ S+S +G+S +C+A A L L++ACG G +C+AIQPG CY
Sbjct: 340 TSVYPLSLSGGSSSAALNGSSMFCVAKADADDDKLVDGLNWACGQGRANCAAIQPGQPCY 399
Query: 64 NPNSVHDHASFAFNKYYQK-NPVPNSCNFGGNAVLTNTNPSKASTIY 109
PN V HASFAFN YYQK +C+F G A+ T +PS + Y
Sbjct: 400 LPNDVKSHASFAFNDYYQKMKSAGGTCDFDGTAITTTRDPSYRTCAY 446
>E133_ARATH (Q9ZU91) Putative glucan endo-1,3-beta-glucosidase 3
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta-glucanase)
(Beta-1,3-endoglucanase) (Beta-1,3-glucanase)
Length = 501
Score = 87.4 bits (215), Expect = 5e-18
Identities = 39/90 (43%), Positives = 58/90 (64%), Gaps = 1/90 (1%)
Query: 21 SSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYY 80
++ ++CIA ++ LQ ALD+ACG G DCSA+ G SCY P+ V H+++AFN YY
Sbjct: 355 TTNQTFCIAKEKVDRKMLQAALDWACGPGKVDCSALMQGESCYEPDDVVAHSTYAFNAYY 414
Query: 81 QK-NPVPNSCNFGGNAVLTNTNPSKASTIY 109
QK SC+F G A +T T+PS+ + ++
Sbjct: 415 QKMGKASGSCDFKGVATVTTTDPSRGTCVF 444
>E13B_WHEAT (P52409) Glucan endo-1,3-beta-glucosidase precursor (EC
3.2.1.39) ((1->3)-beta-glucan endohydrolase)
((1->3)-beta-glucanase) (Beta-1,3-endoglucanase)
Length = 461
Score = 85.9 bits (211), Expect = 1e-17
Identities = 42/111 (37%), Positives = 64/111 (56%), Gaps = 8/111 (7%)
Query: 5 SSTVPITAPSTSNSPV-----SSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPG 59
+S P +P+ S +P S G WC+A A+ LQ ++YACG+ DC IQ G
Sbjct: 351 ASVAPTPSPNPSPNPSPKPAPSGGGKWCVAKDGANGTDLQNNINYACGF--VDCKPIQSG 408
Query: 60 GSCYNPNSVHDHASFAFNKYYQKNPVPN-SCNFGGNAVLTNTNPSKASTIY 109
G+C++PNS+ HAS+ N YYQ N + +C+F G ++T+++PS Y
Sbjct: 409 GACFSPNSLQAHASYVMNAYYQANGHTDLACDFKGTGIVTSSDPSYGGCKY 459
>EA6_ARATH (Q06915) Probable glucan endo-1,3-beta-glucosidase A6
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta-glucanase)
(Beta-1,3-endoglucanase) (Anther-specific protein A6)
Length = 478
Score = 67.8 bits (164), Expect = 4e-12
Identities = 34/91 (37%), Positives = 46/91 (50%), Gaps = 1/91 (1%)
Query: 13 PSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYNPNSVHDHA 72
P +N+ G WC+ A++ L+ L AC T C+A+ PG CY P S++ HA
Sbjct: 376 PKPTNNVPYKGQVWCVPVEGANETELEETLRMACAQSNTTCAALAPGRECYEPVSIYWHA 435
Query: 73 SFAFNKYY-QKNPVPNSCNFGGNAVLTNTNP 102
S+A N Y+ Q C F G A T TNP
Sbjct: 436 SYALNSYWAQFRNQSIQCFFNGLAHETTTNP 466
>E135_ARATH (Q9M088) Putative glucan endo-1,3-beta-glucosidase 5
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta-glucanase)
(Beta-1,3-endoglucanase) (Beta-1,3-glucanase)
Length = 484
Score = 56.6 bits (135), Expect = 1e-08
Identities = 30/84 (35%), Positives = 52/84 (61%), Gaps = 6/84 (7%)
Query: 26 WCIASPSASQRSLQV--ALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQ-K 82
WCI +P+A+ + Q+ ++ YAC + DC+++ G SC N N + + S+AFN YYQ
Sbjct: 365 WCILAPNANLQDPQLGPSVSYACDH--ADCTSLGYGSSCGNLN-LAQNVSYAFNSYYQVS 421
Query: 83 NPVPNSCNFGGNAVLTNTNPSKAS 106
N + ++C F G ++++ +PS S
Sbjct: 422 NQLDSACKFPGLSIVSTRDPSVGS 445
>E136_ARATH (Q93Z08) Putative glucan endo-1,3-beta-glucosidase 6
precursor (EC 3.2.1.39) ((1->3)-beta-glucan
endohydrolase) ((1->3)-beta-glucanase)
(Beta-1,3-endoglucanase) (Beta-1,3-glucanase)
Length = 477
Score = 50.8 bits (120), Expect = 5e-07
Identities = 29/81 (35%), Positives = 45/81 (54%), Gaps = 6/81 (7%)
Query: 26 WCIASPSASQRSLQVA--LDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQ-K 82
WC+ P+ QVA + YAC G DC+++ G SC N + + S+AFN YYQ +
Sbjct: 362 WCVMKPNVRLDDPQVAPAVSYACSLG--DCTSLGVGTSCANLDG-KQNISYAFNSYYQIQ 418
Query: 83 NPVPNSCNFGGNAVLTNTNPS 103
+ + +C F + +T T+PS
Sbjct: 419 DQLDTACKFPNISEVTKTDPS 439
>EPD1_CANMA (P56092) EPD1 protein precursor (Essential for
pseudohyphal development 1)
Length = 549
Score = 42.4 bits (98), Expect = 2e-04
Identities = 29/114 (25%), Positives = 48/114 (41%), Gaps = 9/114 (7%)
Query: 2 YSPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGS 61
+ S+ +P T P S + C+ + + S DY C DCS I +
Sbjct: 354 WEASTELPPT-PDKDICDCMSSSLKCVVADNVSTDDYSDLFDYVCAK--IDCSGINANAT 410
Query: 62 -----CYNPNSVHDHASFAFNKYY-QKNPVPNSCNFGGNAVLTNTNPSKASTIY 109
Y+P D SF N YY ++N ++C+F G+A L + + + + Y
Sbjct: 411 TGDYGAYSPCGAKDKLSFVLNLYYEEQNESKSACDFSGSASLQSASTASSCAAY 464
>PHR2_CANAL (O13318) pH responsive protein 2 precursor (pH-regulated
protein 2)
Length = 546
Score = 40.8 bits (94), Expect = 5e-04
Identities = 29/114 (25%), Positives = 44/114 (38%), Gaps = 9/114 (7%)
Query: 2 YSPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGS 61
+ S+ +P T P S + C+ Y C DC I G+
Sbjct: 356 WEASTNLPPT-PDKEVCECMSASLKCVVDDKVDSDDYSDLFSYICAK--IDCDGINANGT 412
Query: 62 -----CYNPNSVHDHASFAFNKYYQKNPVPNS-CNFGGNAVLTNTNPSKASTIY 109
Y+P D SF N YY++N S C+FGG+A L + + + + Y
Sbjct: 413 TGEYGAYSPCHSKDKLSFVMNLYYEQNKESKSACDFGGSASLQSAKTASSCSAY 466
>EPD2_CANMA (O74137) EPD2 protein precursor (Essential for
pseudohyphal development 2)
Length = 549
Score = 35.0 bits (79), Expect = 0.030
Identities = 29/123 (23%), Positives = 49/123 (39%), Gaps = 9/123 (7%)
Query: 1 TYSPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGG 60
T+ S+ +P T +S + C+ + CGY DC I G
Sbjct: 365 TWRASTELPPTPDEEFCDCISQSFN-CVVADDVDAEDYSTLFGEVCGY--IDCGDISANG 421
Query: 61 SC-----YNPNSVHDHASFAFNKYYQ-KNPVPNSCNFGGNAVLTNTNPSKASTIYDHGFK 114
+ ++ S D S+ N+YY +N ++C+F G+A + + + S G
Sbjct: 422 NTGEYGGFSFCSDKDRLSYVLNQYYHDQNERADACDFAGSASINDNASASTSCSAAGGRG 481
Query: 115 LQS 117
LQS
Sbjct: 482 LQS 484
>FIG2_YEAST (P25653) Factor induced gene 2 precursor (Cell wall
adhesin FIG2)
Length = 1609
Score = 31.2 bits (69), Expect = 0.43
Identities = 29/107 (27%), Positives = 50/107 (46%), Gaps = 7/107 (6%)
Query: 3 SPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPGGSC 62
S S +P+TAPS+S+ P S+ ++ +PS S S A D ++ + GS
Sbjct: 145 SSSFELPVTAPSSSSLPSSTSLTFTSVNPSQSWTSFNSEKSSALS-STIDFTSSEISGST 203
Query: 63 YNPNSVH--DHASFAFNKYYQKNPVPNSCNFGGNAVLTNTNPSKAST 107
+P S+ D + Y +P P+S N ++L+ P +S+
Sbjct: 204 -SPKSLESFDTTGTITSSY---SPSPSSKNSNQTSLLSPLEPLSSSS 246
>YNR6_YEAST (P53882) Hypothetical 67.4 kDa protein in RPS3-PSD1
intergenic region
Length = 636
Score = 30.8 bits (68), Expect = 0.57
Identities = 17/63 (26%), Positives = 33/63 (51%), Gaps = 1/63 (1%)
Query: 6 STVPITAPSTSNSPVSSGASWC-IASPSASQRSLQVALDYACGYGGTDCSAIQPGGSCYN 64
S++ T P+ S+S S +SW ++SPS+ S ++L + Y T + S ++
Sbjct: 141 SSLTSTTPTLSSSATSLSSSWSSLSSPSSLLVSSSLSLSLSSSYSDTKLFSFDSRSSIFS 200
Query: 65 PNS 67
P++
Sbjct: 201 PST 203
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 29.6 bits (65), Expect = 1.3
Identities = 29/109 (26%), Positives = 47/109 (42%), Gaps = 5/109 (4%)
Query: 1 TYSPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSL-QVALDYACGYGGTDCSAIQPG 59
T + SS+ ++ STS SP S+ S + S S+S S Q + + T S+
Sbjct: 210 TSTSSSSTSTSSSSTSTSPSSTSTSSSLTSTSSSSTSTSQSSTSTSSSSTSTSPSSTSTS 269
Query: 60 GS--CYNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAVLTNTNPSKAS 106
S +P+S AS Y + P+ + + L +T+PS S
Sbjct: 270 SSSTSTSPSSKSTSASSTSTSSYSTSTSPSLTS--SSPTLASTSPSSTS 316
>ADG3_SCHPO (O74851) Adg3 protein precursor
Length = 1131
Score = 29.6 bits (65), Expect = 1.3
Identities = 29/108 (26%), Positives = 45/108 (40%), Gaps = 11/108 (10%)
Query: 14 STSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQ---PGGSCYNPNSVHD 70
+TS S ++SG S +ASP++S + L + G S+ P S S+H
Sbjct: 404 ATSKSGLNSGVSTLLASPTSSSTFVTSLLRRSSIDGSASSSSASLAVPTVSSSTTGSLHY 463
Query: 71 HASF------AFNKYY--QKNPVPNSCNFGGNAVLTNTNPSKASTIYD 110
+ F +Y PV +S F T+T+ +S IYD
Sbjct: 464 KTTTTVWVTEVFTRYLGDDSTPVTSSSIFSTATEATDTSVQTSSAIYD 511
>SIM1_YEAST (P40472) SIM1 protein precursor
Length = 475
Score = 29.3 bits (64), Expect = 1.6
Identities = 19/43 (44%), Positives = 24/43 (55%)
Query: 3 SPSSTVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYA 45
S SST P T+ STS S SS +S +S S+S S + D A
Sbjct: 167 STSSTSPSTSTSTSTSSTSSSSSSSSSSSSSSSGSGSIYGDLA 209
>ZCH2_HUMAN (Q9C0B9) Zinc finger CCHC domain containing protein 2
(Fragment)
Length = 899
Score = 28.9 bits (63), Expect = 2.2
Identities = 22/85 (25%), Positives = 33/85 (37%), Gaps = 4/85 (4%)
Query: 6 STVPITAPSTSNSPVSSGASWCIASPSASQRSLQVALDYACGYGGTDCSAIQPG----GS 61
+TVP P+ + P S + S + S + ++ GT Q G GS
Sbjct: 676 ATVPPAVPTHTPGPAPSPSPALTHSTAQSDSTSYISAVGNTNANGTVVPPQQMGSGPCGS 735
Query: 62 CYNPNSVHDHASFAFNKYYQKNPVP 86
C S + + N YY NP+P
Sbjct: 736 CGRRCSCGTNGNLQLNSYYYPNPMP 760
>HRG_HUMAN (P04196) Histidine-rich glycoprotein precursor
(Histidine-proline rich glycoprotein) (HPRG)
Length = 525
Score = 28.9 bits (63), Expect = 2.2
Identities = 10/21 (47%), Positives = 14/21 (66%)
Query: 38 LQVALDYACGYGGTDCSAIQP 58
L + L Y+C TDCSA++P
Sbjct: 9 LLITLQYSCAVSPTDCSAVEP 29
>R1AB_IBVBC (Q91QT2) Replicase polyprotein 1ab (pp1ab) (ORF1ab
polyprotein) [Includes: Replicase polyprotein 1a (pp1a)
(ORF1a)] [Contains: p87; p195 (EC 3.4.24.-) (Papain-like
proteinase) (PL-PRO); Peptide HD2 (p41); 3C-like
proteinase (EC 3.4.24.-) (3CL-
Length = 6629
Score = 28.5 bits (62), Expect = 2.8
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Query: 41 ALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAV 96
A+D A Y + QP G+C +VH+ + FA + +P P+ ++GG +V
Sbjct: 3804 AVDPADTYCKYVAAGNQPLGNCVKMLTVHNGSGFAITS--KPSPTPDQDSYGGASV 3857
>R1AB_IBVB (P27920) Replicase polyprotein 1ab (pp1ab) (ORF1ab
polyprotein) [Includes: Replicase polyprotein 1a (pp1a)
(ORF1a)] [Contains: p87; p195 (EC 3.4.24.-) (Papain-like
proteinase) (PL-PRO); Peptide HD2 (p41); 3C-like
proteinase (EC 3.4.24.-) (3CL-P
Length = 6629
Score = 28.5 bits (62), Expect = 2.8
Identities = 17/56 (30%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Query: 41 ALDYACGYGGTDCSAIQPGGSCYNPNSVHDHASFAFNKYYQKNPVPNSCNFGGNAV 96
A+D A Y + QP G+C +VH+ + FA + +P P+ ++GG +V
Sbjct: 3804 AVDPADTYCKYVAAGNQPLGNCVKMLTVHNGSGFAITS--KPSPTPDQDSYGGASV 3857
>LMA1_MOUSE (P19137) Laminin alpha-1 chain precursor (Laminin A
chain)
Length = 3084
Score = 28.5 bits (62), Expect = 2.8
Identities = 20/58 (34%), Positives = 26/58 (44%), Gaps = 12/58 (20%)
Query: 28 IASPSASQRSLQVALDYA-C--GYGGTDCSAIQPG---------GSCYNPNSVHDHAS 73
IASP+A ++ +++ C GY GT C A PG G P H HAS
Sbjct: 699 IASPNAIDLAVAADVEHCECPQGYTGTSCEACLPGYYRVDGILFGGICQPCECHGHAS 756
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.310 0.125 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,337,770
Number of Sequences: 164201
Number of extensions: 699697
Number of successful extensions: 1527
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 1472
Number of HSP's gapped (non-prelim): 66
length of query: 120
length of database: 59,974,054
effective HSP length: 96
effective length of query: 24
effective length of database: 44,210,758
effective search space: 1061058192
effective search space used: 1061058192
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147496.8


