

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147483.4 + phase: 0
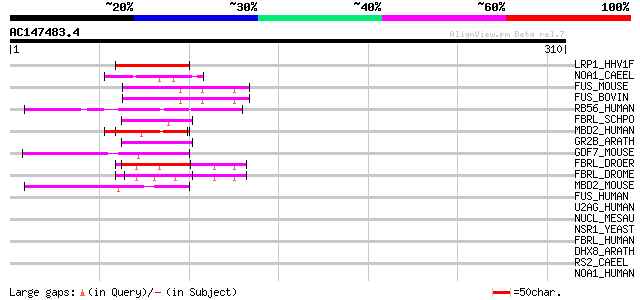
(310 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LRP1_HHV1F (P17588) Latency-related protein 1 50 7e-06
NOA1_CAEEL (Q9TYK1) Putative nucleolar protein family A member 1... 50 9e-06
FUS_MOUSE (P56959) RNA-binding protein FUS (Pigpen protein) 47 8e-05
FUS_BOVIN (Q28009) RNA-binding protein FUS (Pigpen protein) 47 8e-05
RB56_HUMAN (Q92804) TATA-binding protein associated factor 2N (R... 46 1e-04
FBRL_SCHPO (P35551) Fibrillarin 46 1e-04
MBD2_HUMAN (Q9UBB5) Methyl-CpG binding protein 2 (Methyl-CpG bin... 45 2e-04
GR2B_ARATH (Q38896) Glycine-rich protein 2b (AtGRP2b) 44 4e-04
GDF7_MOUSE (P43029) Growth/differentiation factor 7 precursor (G... 44 5e-04
FBRL_DROER (Q8I1F4) Fibrillarin 44 5e-04
FBRL_DROME (Q9W1V3) Fibrillarin 44 7e-04
MBD2_MOUSE (Q9Z2E1) Methyl-CpG binding protein 2 (Methyl-CpG bin... 43 9e-04
FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncog... 42 0.001
U2AG_HUMAN (Q01081) Splicing factor U2AF 35 kDa subunit (U2 auxi... 42 0.002
NUCL_MESAU (P08199) Nucleolin (Protein C23) 42 0.002
NSR1_YEAST (P27476) Nuclear localization sequence binding protei... 42 0.002
FBRL_HUMAN (P22087) Fibrillarin (34 kDa nucleolar scleroderma an... 42 0.002
DHX8_ARATH (Q38953) Putative pre-mRNA splicing factor ATP-depend... 42 0.002
RS2_CAEEL (P51403) 40S ribosomal protein S2 42 0.003
NOA1_HUMAN (Q9NY12) Nucleolar protein family A member 1 (snoRNP ... 42 0.003
>LRP1_HHV1F (P17588) Latency-related protein 1
Length = 340
Score = 50.1 bits (118), Expect = 7e-06
Identities = 25/41 (60%), Positives = 26/41 (62%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
R G G GRG G GRG +RGRG RGRGGRGG GR G
Sbjct: 288 RGGGSGGGRGPGGGRGGPRGSRGRGGRGRGGRGGGRRGRQG 328
>NOA1_CAEEL (Q9TYK1) Putative nucleolar protein family A member 1
(snoRNP protein GAR1) (H/ACA ribonucleoprotein GAR1)
Length = 244
Score = 49.7 bits (117), Expect = 9e-06
Identities = 34/68 (50%), Positives = 38/68 (55%), Gaps = 16/68 (23%)
Query: 54 PMNRFVRDDGDGSGRGRGRGRGRDVYARG-----RGDRGRGG--------RGGRGDGRGG 100
P++RF+ G G GRG GRGRG D RG RG GRGG RGG G GRGG
Sbjct: 141 PVDRFLPQAGGGRGRG-GRGRGGDRGGRGSDRGGRGGFGRGGGGGFRGGDRGGFGGGRGG 199
Query: 101 FKRYGDDR 108
F+ G DR
Sbjct: 200 FR--GGDR 205
Score = 37.4 bits (85), Expect = 0.048
Identities = 24/55 (43%), Positives = 28/55 (50%), Gaps = 1/55 (1%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDI 114
R G G GRG G G GDRG G RGGRG G GG R G D+ +++
Sbjct: 7 RGGGGGFRGGRGGGGGGGFRGGRGGDRGGGFRGGRG-GFGGGGRGGYDQGPPEEV 60
Score = 33.1 bits (74), Expect = 0.90
Identities = 21/38 (55%), Positives = 22/38 (57%), Gaps = 4/38 (10%)
Query: 69 GRGRGRGRDVYARGRGDRGRGG-RGGRGDGRGGFKRYG 105
GRG G G + GRG G GG RGGRG RGG R G
Sbjct: 6 GRGGGGG---FRGGRGGGGGGGFRGGRGGDRGGGFRGG 40
>FUS_MOUSE (P56959) RNA-binding protein FUS (Pigpen protein)
Length = 518
Score = 46.6 bits (109), Expect = 8e-05
Identities = 33/83 (39%), Positives = 43/83 (51%), Gaps = 12/83 (14%)
Query: 64 DGSGRGRGRGRGRDVYARGRGDRGR-GGRGGR----GDGRGGFKRYGD--DRTSHQDIAR 116
D GRGRG G G + + G RGR GGRGGR G RGGF ++G D+ S D +
Sbjct: 213 DRGGRGRGGGGGYNRSSGGYEPRGRGGGRGGRGGMGGSDRGGFNKFGGPRDQGSRHDSEQ 272
Query: 117 SNADGLYV-----GDNADGEKLA 134
N+D + G+N E +A
Sbjct: 273 DNSDNNTIFVQGLGENVTIESVA 295
Score = 39.3 bits (90), Expect = 0.013
Identities = 27/53 (50%), Positives = 29/53 (53%), Gaps = 6/53 (11%)
Query: 58 FVRDDGDG-SGRGRGRGRGRDVYARGRGDRGRGGRGG---RGDGRGGFKRYGD 106
F R G+G GRGRG GR Y G G G GGRGG G G GG +R GD
Sbjct: 368 FNRGGGNGRGGRGRGGPMGRGGY--GGGGSGGGGRGGFPSGGGGGGGQQRAGD 418
Score = 38.9 bits (89), Expect = 0.016
Identities = 28/55 (50%), Positives = 30/55 (53%), Gaps = 11/55 (20%)
Query: 51 RRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
RR NR G G+GRG GRGRG + GRG G GG GG GRGGF G
Sbjct: 364 RRADFNR-----GGGNGRG-GRGRGGPM---GRGGYGGGGSGG--GGRGGFPSGG 407
Score = 38.5 bits (88), Expect = 0.021
Identities = 28/69 (40%), Positives = 31/69 (44%), Gaps = 5/69 (7%)
Query: 45 ESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYA---RGRGDRGRGGRGGRGDGRGGF 101
E+ N R N+ DG G G G Y RGRG RGG GRG RGGF
Sbjct: 427 ENMNFSWRNECNQCKAPKPDGPGGGPGGSHMGGNYGDDRRGRGGYDRGGYRGRGGDRGGF 486
Query: 102 K--RYGDDR 108
+ R G DR
Sbjct: 487 RGGRGGGDR 495
Score = 30.0 bits (66), Expect = 7.6
Identities = 31/104 (29%), Positives = 35/104 (32%), Gaps = 18/104 (17%)
Query: 8 PGGGRGRGKPLEEAAQEAPQAPVVNRHIRVR------QTPADAESDNVPRRQPMNRFVRD 61
P GG G G + P N + R + P P M D
Sbjct: 404 PSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNECNQCKAPKPDGPGGGPGGSHMGGNYGD 463
Query: 62 DGDGSG-------RGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
D G G RGRG RG RG RG G RGG G G+
Sbjct: 464 DRRGRGGYDRGGYRGRGGDRG-----GFRGGRGGGDRGGFGPGK 502
>FUS_BOVIN (Q28009) RNA-binding protein FUS (Pigpen protein)
Length = 512
Score = 46.6 bits (109), Expect = 8e-05
Identities = 33/83 (39%), Positives = 43/83 (51%), Gaps = 12/83 (14%)
Query: 64 DGSGRGRGRGRGRDVYARGRGDRGR-GGRGGR----GDGRGGFKRYGD--DRTSHQDIAR 116
D GRGRG G G + + G RGR GGRGGR G RGGF ++G D+ S D +
Sbjct: 206 DRGGRGRGGGGGYNRSSGGYEPRGRGGGRGGRGGMGGSDRGGFNKFGGPRDQGSRHDSEQ 265
Query: 117 SNADGLYV-----GDNADGEKLA 134
N+D + G+N E +A
Sbjct: 266 DNSDNNTIFVQGLGENVTIESVA 288
Score = 39.3 bits (90), Expect = 0.013
Identities = 27/53 (50%), Positives = 29/53 (53%), Gaps = 6/53 (11%)
Query: 58 FVRDDGDG-SGRGRGRGRGRDVYARGRGDRGRGGRGG---RGDGRGGFKRYGD 106
F R G+G GRGRG GR Y G G G GGRGG G G GG +R GD
Sbjct: 361 FNRGGGNGRGGRGRGGPMGRGGY--GGGGSGGGGRGGFPSGGGGGGGQQRAGD 411
Score = 38.9 bits (89), Expect = 0.016
Identities = 28/55 (50%), Positives = 30/55 (53%), Gaps = 11/55 (20%)
Query: 51 RRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
RR NR G G+GRG GRGRG + GRG G GG GG GRGGF G
Sbjct: 357 RRADFNR-----GGGNGRG-GRGRGGPM---GRGGYGGGGSGG--GGRGGFPSGG 400
Score = 36.6 bits (83), Expect = 0.082
Identities = 24/53 (45%), Positives = 26/53 (48%), Gaps = 4/53 (7%)
Query: 60 RDDGDGSGRGRGRGRGR--DVYARGRGDRGRGGRGGRGDGRGGFK--RYGDDR 108
+ DG G G G G D GRG RGG GRG RGGF+ R G DR
Sbjct: 437 KPDGPGGGPGGSHMGGNYGDDRRGGRGGYDRGGYRGRGGDRGGFRGGRGGGDR 489
Score = 35.0 bits (79), Expect = 0.24
Identities = 34/90 (37%), Positives = 37/90 (40%), Gaps = 19/90 (21%)
Query: 45 ESDNVPRRQPMNRFVRDDGDGSGRGRG-----------RGRGRDVYARGRGDRGRGG-RG 92
E+ N R N+ DG G G G R GR Y RG G RGRGG RG
Sbjct: 420 ENMNFSWRNECNQCKAPKPDGPGGGPGGSHMGGNYGDDRRGGRGGYDRG-GYRGRGGDRG 478
Query: 93 GRGDGRGGFKRYG------DDRTSHQDIAR 116
G GRGG R G D R H+ R
Sbjct: 479 GFRGGRGGGDRGGFGPGKMDSRGEHRQDRR 508
>RB56_HUMAN (Q92804) TATA-binding protein associated factor 2N
(RNA-binding protein 56) (TAFII68) (TAF(II)68)
Length = 592
Score = 45.8 bits (107), Expect = 1e-04
Identities = 42/123 (34%), Positives = 52/123 (42%), Gaps = 14/123 (11%)
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDGSGR 68
GG +GRG + P N + R + N PR + D G
Sbjct: 344 GGFQGRGGDPKSGDWVCPNPSCGNMNFARRNS---CNQCNEPRPE-------DSRPSGGD 393
Query: 69 GRGRGRGRDVYARGRGDRGRGG-RGGRGDGRGGFKRYGDDRTSHQDIARSNADGLYVGDN 127
RGRG G + RGRG GRGG RGG G R G YG DR+S + + G Y GD
Sbjct: 394 FRGRGYGGERGYRGRG--GRGGDRGGYGGDRSG-GGYGGDRSSGGGYSGDRSGGGYGGDR 450
Query: 128 ADG 130
+ G
Sbjct: 451 SGG 453
Score = 41.6 bits (96), Expect = 0.003
Identities = 36/105 (34%), Positives = 55/105 (52%), Gaps = 15/105 (14%)
Query: 52 RQPMNRFVRDDGDGSGRGRGRGRGRDVYAR-GRGDRGRGGRGGRGDGRGGFKRYGDDRTS 110
R+ ++R+ +D G G +G GRGR Y + GRG G G RGGFK +G
Sbjct: 164 RRDVSRY-GEDNRGYGGSQGGGRGRGGYDKDGRGPM----TGSSGGDRGGFKNFG----G 214
Query: 111 HQDIA-RSNADGLYVGDNADGEKL-AKKLGPEI-MDQITEAYEEI 152
H+D R++AD DN+D + + LG + DQ+ E +++I
Sbjct: 215 HRDYGPRTDADS--ESDNSDNNTIFVQGLGEGVSTDQVGEFFKQI 257
Score = 39.3 bits (90), Expect = 0.013
Identities = 34/83 (40%), Positives = 35/83 (41%), Gaps = 17/83 (20%)
Query: 63 GDGSGRGRGRGRGRDVYARGRG----DRG-----RGG------RGGRGDGRGGFKRYGDD 107
G G RG G G R Y RG DRG RGG RGG G RGG YG D
Sbjct: 485 GYGGDRGGGYGGDRGGYGGDRGGYGGDRGGYGGDRGGYGGDRSRGGYGGDRGGGSGYGGD 544
Query: 108 RTSHQDIARSNADGLYVGDNADG 130
R+ RS G Y GD G
Sbjct: 545 RSGGYGGDRSG--GGYGGDRGGG 565
Score = 38.9 bits (89), Expect = 0.016
Identities = 28/75 (37%), Positives = 31/75 (41%), Gaps = 7/75 (9%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGF----KRYGDDRTSHQDIA 115
R G G RG G G R Y RG G RGG G RGG+ YG DR +
Sbjct: 467 RGGGYGGDRGGGYGGDRGGYGGDRGGGYGGDRGGYGGDRGGYGGDRGGYGGDRGGY---G 523
Query: 116 RSNADGLYVGDNADG 130
+ G Y GD G
Sbjct: 524 GDRSRGGYGGDRGGG 538
Score = 34.7 bits (78), Expect = 0.31
Identities = 24/52 (46%), Positives = 25/52 (47%), Gaps = 6/52 (11%)
Query: 63 GDGSGRGRGRGRGRDVYARGRG-----DRGRGGRGGRGDGRGGFK-RYGDDR 108
GD SG G G R Y RG DRG G G RG G GG + YG DR
Sbjct: 439 GDRSGGGYGGDRSGGGYGGDRGGGYGGDRGGGYGGDRGGGYGGDRGGYGGDR 490
Score = 33.5 bits (75), Expect = 0.69
Identities = 20/39 (51%), Positives = 20/39 (51%), Gaps = 1/39 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGG-RGDGRGG 100
G G RG G G G D GDR GG GG RG G GG
Sbjct: 530 GYGGDRGGGSGYGGDRSGGYGGDRSGGGYGGDRGGGYGG 568
Score = 30.8 bits (68), Expect = 4.5
Identities = 20/41 (48%), Positives = 21/41 (50%), Gaps = 11/41 (26%)
Query: 68 RGRGRGRGRDVYARGRGDRGRGGRGGRG--DGRGGFKRYGD 106
RG G G GR RGRGG GRG GRGG + GD
Sbjct: 326 RGGGSGGGR---------RGRGGYRGRGGFQGRGGDPKSGD 357
Score = 30.4 bits (67), Expect = 5.8
Identities = 24/54 (44%), Positives = 26/54 (47%), Gaps = 6/54 (11%)
Query: 61 DDGDGSGRG--RGRGRGRDVYARGRG-DRGRG---GRGGRGDGRGGFKRYGDDR 108
D G GSG G R G G D G G DRG G RGG G GG Y +D+
Sbjct: 534 DRGGGSGYGGDRSGGYGGDRSGGGYGGDRGGGYGGDRGGYGGKMGGRNDYRNDQ 587
Score = 29.6 bits (65), Expect = 10.0
Identities = 29/74 (39%), Positives = 30/74 (40%), Gaps = 10/74 (13%)
Query: 63 GDGSGRGRGRGRGRDVYARG---RGDRGRGGRGG-RGDGRGGFK--RYGDDRTSHQDIAR 116
G G G R G G G GDR GG GG RG G GG + YG DR R
Sbjct: 424 GGGYGGDRSSGGGYSGDRSGGGYGGDRSGGGYGGDRGGGYGGDRGGGYGGDRGGGYGGDR 483
Query: 117 SNADGLYVGDNADG 130
G Y GD G
Sbjct: 484 ----GGYGGDRGGG 493
>FBRL_SCHPO (P35551) Fibrillarin
Length = 305
Score = 45.8 bits (107), Expect = 1e-04
Identities = 25/42 (59%), Positives = 25/42 (59%), Gaps = 2/42 (4%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRG--RGGRGGRGDGRGGFK 102
G G GRG RG GR GRG RG RGGRGG GRGG K
Sbjct: 27 GFGGGRGGARGGGRGGARGGRGGRGGARGGRGGSSGGRGGAK 68
Score = 34.7 bits (78), Expect = 0.31
Identities = 20/44 (45%), Positives = 21/44 (47%), Gaps = 6/44 (13%)
Query: 63 GDGSGRGRGRGR------GRDVYARGRGDRGRGGRGGRGDGRGG 100
G GRG RG GR + GRG GGRGG GRGG
Sbjct: 6 GSRGGRGGSRGGRGGFNGGRGGFGGGRGGARGGGRGGARGGRGG 49
Score = 32.0 bits (71), Expect = 2.0
Identities = 21/41 (51%), Positives = 22/41 (53%), Gaps = 7/41 (17%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
GS GRG RG GRG GGRGG G GRGG + G
Sbjct: 6 GSRGGRGGSRG------GRGGFN-GGRGGFGGGRGGARGGG 39
>MBD2_HUMAN (Q9UBB5) Methyl-CpG binding protein 2 (Methyl-CpG
binding domain protein 2) (Demethylase) (DMTase)
Length = 411
Score = 45.1 bits (105), Expect = 2e-04
Identities = 27/50 (54%), Positives = 30/50 (60%), Gaps = 5/50 (10%)
Query: 54 PMNRFVRDDGDGSGRGRGR----GRGRDVYARGRGDRGRGGRGGRGDGRG 99
P++ R+ G GRGRGR GRG V RGRG RGRG GRG GRG
Sbjct: 45 PVSGVRREGARGGGRGRGRWKQAGRGGGVCGRGRG-RGRGRGRGRGRGRG 93
Score = 43.9 bits (102), Expect = 5e-04
Identities = 24/41 (58%), Positives = 24/41 (58%), Gaps = 3/41 (7%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
R G G GRGRGRGRGR RGR G G GG G G GG
Sbjct: 76 RGRGRGRGRGRGRGRGR---GRGRPPSGGSGLGGDGGGCGG 113
Score = 30.4 bits (67), Expect = 5.8
Identities = 20/44 (45%), Positives = 20/44 (45%), Gaps = 4/44 (9%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKR 103
R G G GRGRGR G G GG GG G G GG R
Sbjct: 84 RGRGRGRGRGRGRPPSGGSGLGGDG----GGCGGGGSGGGGAPR 123
>GR2B_ARATH (Q38896) Glycine-rich protein 2b (AtGRP2b)
Length = 201
Score = 44.3 bits (103), Expect = 4e-04
Identities = 22/40 (55%), Positives = 22/40 (55%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFK 102
G G G GRG GRG Y G G RG GGRGG G FK
Sbjct: 101 GFGGGGGRGGGRGGGSYGGGYGGRGSGGRGGGGGDNSCFK 140
Score = 31.6 bits (70), Expect = 2.6
Identities = 23/58 (39%), Positives = 26/58 (44%), Gaps = 7/58 (12%)
Query: 43 DAESDNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
D E DN R + + D G G G + GRG G GG GGRG GRGG
Sbjct: 64 DVEVDNSGRPKAIEVSGPDGAPVQGNSGGGGS-----SGGRG--GFGGGGGRGGGRGG 114
>GDF7_MOUSE (P43029) Growth/differentiation factor 7 precursor
(GDF-7)
Length = 461
Score = 43.9 bits (102), Expect = 5e-04
Identities = 33/97 (34%), Positives = 40/97 (41%), Gaps = 9/97 (9%)
Query: 8 PGGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRDDGDGSG 67
PGGG G G EE A + + R+ A A + P D G G+G
Sbjct: 248 PGGGDGGGTAAEERALLVISSRTQRKESLFREIRAQARALRAAAEPPP-----DPGPGAG 302
Query: 68 RGR----GRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
+ GR R R A RG +G GG GG G G GG
Sbjct: 303 SRKANLGGRRRRRTALAGTRGAQGSGGGGGGGGGGGG 339
>FBRL_DROER (Q8I1F4) Fibrillarin
Length = 345
Score = 43.9 bits (102), Expect = 5e-04
Identities = 26/42 (61%), Positives = 26/42 (61%), Gaps = 3/42 (7%)
Query: 63 GDGSGRGRGRGRGRDVYARG--RGDRGRGGR-GGRGDGRGGF 101
G G G GRGRG G D RG G RG GGR GG G GRGGF
Sbjct: 34 GGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGGGGGRGGF 75
Score = 43.5 bits (101), Expect = 7e-04
Identities = 34/84 (40%), Positives = 36/84 (42%), Gaps = 11/84 (13%)
Query: 60 RDDGDGSGRG----RGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQD-- 113
R G G GRG RG G GR RG G RG GGRGG G G K + H+
Sbjct: 64 RGGGGGGGRGGFGGRGGGGGRGGGGRGGGGRGGGGRGGGAGGFKGGKTVTIEPHRHEGVF 123
Query: 114 IARSNADGLYV-----GDNADGEK 132
IAR D L G GEK
Sbjct: 124 IARGKEDALVTRNFVPGSEVYGEK 147
Score = 42.4 bits (98), Expect = 0.001
Identities = 25/47 (53%), Positives = 26/47 (55%), Gaps = 9/47 (19%)
Query: 65 GSGRGRGRGRGRDVY-------ARGRGDRGRGGR--GGRGDGRGGFK 102
G GRG G G GR + RG G RG GGR GGRG G GGFK
Sbjct: 61 GGGRGGGGGGGRGGFGGRGGGGGRGGGGRGGGGRGGGGRGGGAGGFK 107
Score = 40.8 bits (94), Expect = 0.004
Identities = 25/43 (58%), Positives = 25/43 (58%), Gaps = 8/43 (18%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G G G G GRGRG G GD RGGRGG G GRGG R G
Sbjct: 32 GGGGGFGGGRGRG------GGGD--RGGRGGFGGGRGGGGRGG 66
Score = 39.7 bits (91), Expect = 0.010
Identities = 27/59 (45%), Positives = 28/59 (46%), Gaps = 13/59 (22%)
Query: 63 GDGSGRGRGRGR---GRDVYARGRGDRGRGGRGGRG----------DGRGGFKRYGDDR 108
G G GRGRG G GR + GRG GRGG GG G GRGG R G R
Sbjct: 36 GFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGGGGGRGGFGGRGGGGGRGGGGRGGGGR 94
Score = 35.0 bits (79), Expect = 0.24
Identities = 23/51 (45%), Positives = 24/51 (46%), Gaps = 2/51 (3%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDG--RGGFKRYGDDR 108
R G G G G G RGR G G G GG GRG G RGG +G R
Sbjct: 9 RGGGGGGGGGGGGFRGRGGGGGGGGGGGFGGGRGRGGGGDRGGRGGFGGGR 59
>FBRL_DROME (Q9W1V3) Fibrillarin
Length = 344
Score = 43.5 bits (101), Expect = 7e-04
Identities = 25/47 (53%), Positives = 26/47 (55%), Gaps = 9/47 (19%)
Query: 65 GSGRGRGRGRGRDVY-------ARGRGDRGRGGR--GGRGDGRGGFK 102
G GRG G G GR + RG G RG GGR GGRG G GGFK
Sbjct: 60 GGGRGGGGGGGRGAFGGRGGGGGRGGGGRGGGGRGGGGRGGGAGGFK 106
Score = 43.5 bits (101), Expect = 7e-04
Identities = 34/84 (40%), Positives = 36/84 (42%), Gaps = 11/84 (13%)
Query: 60 RDDGDGSGRG----RGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQD-- 113
R G G GRG RG G GR RG G RG GGRGG G G K + H+
Sbjct: 63 RGGGGGGGRGAFGGRGGGGGRGGGGRGGGGRGGGGRGGGAGGFKGGKTVTIEPHRHEGVF 122
Query: 114 IARSNADGLYV-----GDNADGEK 132
IAR D L G GEK
Sbjct: 123 IARGKEDALVTRNFVPGSEVYGEK 146
Score = 41.6 bits (96), Expect = 0.003
Identities = 25/42 (59%), Positives = 25/42 (59%), Gaps = 3/42 (7%)
Query: 63 GDGSGRGRGRGRGRDVYARG--RGDRGRGGR-GGRGDGRGGF 101
G G G GRGRG G D RG G RG GGR GG G GRG F
Sbjct: 33 GGGFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGGGGGRGAF 74
Score = 40.8 bits (94), Expect = 0.004
Identities = 25/43 (58%), Positives = 25/43 (58%), Gaps = 8/43 (18%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G G G G GRGRG G GD RGGRGG G GRGG R G
Sbjct: 31 GGGGGFGGGRGRG------GGGD--RGGRGGFGGGRGGGGRGG 65
Score = 39.7 bits (91), Expect = 0.010
Identities = 27/59 (45%), Positives = 28/59 (46%), Gaps = 13/59 (22%)
Query: 63 GDGSGRGRGRGR---GRDVYARGRGDRGRGGRGGRG----------DGRGGFKRYGDDR 108
G G GRGRG G GR + GRG GRGG GG G GRGG R G R
Sbjct: 35 GFGGGRGRGGGGDRGGRGGFGGGRGGGGRGGGGGGGRGAFGGRGGGGGRGGGGRGGGGR 93
Score = 33.5 bits (75), Expect = 0.69
Identities = 19/35 (54%), Positives = 19/35 (54%), Gaps = 4/35 (11%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRG 99
G G G G G G RGRG G GG GG G GRG
Sbjct: 11 GGGGGGGGGGG----FRGRGGGGGGGGGGFGGGRG 41
Score = 32.7 bits (73), Expect = 1.2
Identities = 23/49 (46%), Positives = 23/49 (46%), Gaps = 9/49 (18%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
R G G G G G RGR G G G GG G G GRGG G DR
Sbjct: 9 RGGGGGGGGGGGGFRGR-----GGGGGGGGGGFGGGRGRGG----GGDR 48
>MBD2_MOUSE (Q9Z2E1) Methyl-CpG binding protein 2 (Methyl-CpG
binding domain protein 2)
Length = 414
Score = 43.1 bits (100), Expect = 9e-04
Identities = 33/94 (35%), Positives = 39/94 (41%), Gaps = 7/94 (7%)
Query: 9 GGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFV--RDDGDGS 66
G G G +E+ Q + AP +R ++ V R G G
Sbjct: 23 GSGAGGDSAIEQGGQGSALAPSPVSGVRREGARGGGRGRGRWKQAARGGGVCGRGRGRGR 82
Query: 67 GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
GRGRGRGRG RGRG GG G GDG GG
Sbjct: 83 GRGRGRGRG-----RGRGRPQSGGSGLGGDGGGG 111
Score = 42.0 bits (97), Expect = 0.002
Identities = 24/44 (54%), Positives = 24/44 (54%), Gaps = 3/44 (6%)
Query: 60 RDDGDGSGRGRGRGRGR---DVYARGRGDRGRGGRGGRGDGRGG 100
R G G GRGRGRGRGR G G G GG GG G G GG
Sbjct: 78 RGRGRGRGRGRGRGRGRGRPQSGGSGLGGDGGGGAGGCGVGSGG 121
Score = 37.7 bits (86), Expect = 0.037
Identities = 24/55 (43%), Positives = 29/55 (52%), Gaps = 3/55 (5%)
Query: 54 PMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
P++ R+ G GRGRGR + ARG G GRG GRG GRG + G R
Sbjct: 45 PVSGVRREGARGGGRGRGRWKQA---ARGGGVCGRGRGRGRGRGRGRGRGRGRGR 96
>FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncogene
TLS) (Translocated in liposarcoma protein) (POMp75) (75
kDa DNA-pairing protein)
Length = 526
Score = 42.4 bits (98), Expect = 0.001
Identities = 32/87 (36%), Positives = 42/87 (47%), Gaps = 12/87 (13%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGR-GGRGGR----GDGRGGFKRYGD--DRTSHQ 112
R G G G G G G + + G RGR GGRGGR G RGGF ++G D+ S
Sbjct: 216 RGRGGSGGGGGGGGGGYNRSSGGYEPRGRGGGRGGRGGMGGSDRGGFNKFGGPRDQGSRH 275
Query: 113 DIARSNADGLYV-----GDNADGEKLA 134
D + N+D + G+N E +A
Sbjct: 276 DSEQDNSDNNTIFVQGLGENVTIESVA 302
Score = 39.3 bits (90), Expect = 0.013
Identities = 27/53 (50%), Positives = 29/53 (53%), Gaps = 6/53 (11%)
Query: 58 FVRDDGDG-SGRGRGRGRGRDVYARGRGDRGRGGRGG---RGDGRGGFKRYGD 106
F R G+G GRGRG GR Y G G G GGRGG G G GG +R GD
Sbjct: 375 FNRGGGNGRGGRGRGGPMGRGGY--GGGGSGGGGRGGFPSGGGGGGGQQRAGD 425
Score = 38.9 bits (89), Expect = 0.016
Identities = 28/55 (50%), Positives = 30/55 (53%), Gaps = 11/55 (20%)
Query: 51 RRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
RR NR G G+GRG GRGRG + GRG G GG GG GRGGF G
Sbjct: 371 RRADFNR-----GGGNGRG-GRGRGGPM---GRGGYGGGGSGG--GGRGGFPSGG 414
Score = 36.6 bits (83), Expect = 0.082
Identities = 24/53 (45%), Positives = 26/53 (48%), Gaps = 4/53 (7%)
Query: 60 RDDGDGSGRGRGRGRGR--DVYARGRGDRGRGGRGGRGDGRGGFK--RYGDDR 108
+ DG G G G G D GRG RGG GRG RGGF+ R G DR
Sbjct: 451 KPDGPGGGPGGSHMGGNYGDDRRGGRGGYDRGGYRGRGGDRGGFRGGRGGGDR 503
Score = 35.8 bits (81), Expect = 0.14
Identities = 22/44 (50%), Positives = 23/44 (52%), Gaps = 6/44 (13%)
Query: 63 GDGSGRGRGR-----GRGRDVYAR-GRGDRGRGGRGGRGDGRGG 100
G GSG G G G G Y + RG RGRGG GG G G GG
Sbjct: 187 GGGSGGGYGNQDQSGGGGSGGYGQQDRGGRGRGGSGGGGGGGGG 230
Score = 35.0 bits (79), Expect = 0.24
Identities = 34/90 (37%), Positives = 37/90 (40%), Gaps = 19/90 (21%)
Query: 45 ESDNVPRRQPMNRFVRDDGDGSGRGRG-----------RGRGRDVYARGRGDRGRGG-RG 92
E+ N R N+ DG G G G R GR Y RG G RGRGG RG
Sbjct: 434 ENMNFSWRNECNQCKAPKPDGPGGGPGGSHMGGNYGDDRRGGRGGYDRG-GYRGRGGDRG 492
Query: 93 GRGDGRGGFKRYG------DDRTSHQDIAR 116
G GRGG R G D R H+ R
Sbjct: 493 GFRGGRGGGDRGGFGPGKMDSRGEHRQDRR 522
>U2AG_HUMAN (Q01081) Splicing factor U2AF 35 kDa subunit (U2
auxiliary factor 35 kDa subunit) (U2 snRNP auxiliary
factor small subunit)
Length = 240
Score = 42.0 bits (97), Expect = 0.002
Identities = 29/71 (40%), Positives = 37/71 (51%), Gaps = 4/71 (5%)
Query: 48 NVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRG-DRGRGGRGGRGDGRGGFKRYGD 106
N +P++R +R + G R + R R R R R DRGRGG GG G G GG +R
Sbjct: 170 NFMHLKPISRELRRELYGRRRKKHRSRSRSRERRSRSRDRGRGGGGGGGGGGGGRER--- 226
Query: 107 DRTSHQDIARS 117
DR +D RS
Sbjct: 227 DRRRSRDRERS 237
>NUCL_MESAU (P08199) Nucleolin (Protein C23)
Length = 713
Score = 42.0 bits (97), Expect = 0.002
Identities = 24/40 (60%), Positives = 24/40 (60%), Gaps = 2/40 (5%)
Query: 63 GDGSGRGR--GRGRGRDVYARGRGDRGRGGRGGRGDGRGG 100
G G GRG GRG GR G G RGRGG GGRG RGG
Sbjct: 654 GRGGGRGGFGGRGGGRGGGRGGFGGRGRGGFGGRGGFRGG 693
Score = 40.8 bits (94), Expect = 0.004
Identities = 24/44 (54%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGD 106
G GRG GRG GR + GRG G GGRGG GRGG GD
Sbjct: 660 GGFGGRGGGRGGGRGGFG-GRGRGGFGGRGGFRGGRGGGGGGGD 702
Score = 37.4 bits (85), Expect = 0.048
Identities = 26/48 (54%), Positives = 26/48 (54%), Gaps = 7/48 (14%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRG--RGGRGGRGDGRGGFKRYG 105
R G G GRG GRGR G G RG RGGRGG G G G FK G
Sbjct: 665 RGGGRGGGRGGFGGRGRG----GFGGRGGFRGGRGG-GGGGGDFKPQG 707
Score = 35.8 bits (81), Expect = 0.14
Identities = 23/51 (45%), Positives = 26/51 (50%), Gaps = 2/51 (3%)
Query: 58 FVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
+ + G+G GRG GRG G GRGG GGRG RGGF G R
Sbjct: 643 WAKPKGEGGFGGRGGGRGGFGGRGGGRGGGRGGFGGRG--RGGFGGRGGFR 691
>NSR1_YEAST (P27476) Nuclear localization sequence binding protein
(P67)
Length = 414
Score = 42.0 bits (97), Expect = 0.002
Identities = 29/59 (49%), Positives = 33/59 (55%), Gaps = 3/59 (5%)
Query: 47 DNVPRRQPMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
DN P R + R + DG GRG RG G RG G+RG GGRGG GRGGF+ G
Sbjct: 334 DNRPVRLDFSS-PRPNNDG-GRGGSRGFGGRGGGRG-GNRGFGGRGGARGGRGGFRPSG 389
>FBRL_HUMAN (P22087) Fibrillarin (34 kDa nucleolar scleroderma
antigen)
Length = 321
Score = 42.0 bits (97), Expect = 0.002
Identities = 25/59 (42%), Positives = 27/59 (45%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADG 121
G G G GRGRG RGRG G GG GG G GG G +R + R N G
Sbjct: 25 GGRGGFGGGRGRGGGFRGRGRGGGGGGGGGGGGGRGGGGFHSGGNRGRGRGGKRGNQSG 83
Score = 36.2 bits (82), Expect = 0.11
Identities = 23/42 (54%), Positives = 23/42 (54%), Gaps = 7/42 (16%)
Query: 63 GDGSGRGRGRGRGRDVYARGRGD----RGRGGRGGRGDGRGG 100
GD GRG GRG RGRG RGRGG GG G G GG
Sbjct: 19 GDRGGRG---GRGGFGGGRGRGGGFRGRGRGGGGGGGGGGGG 57
Score = 32.3 bits (72), Expect = 1.5
Identities = 24/80 (30%), Positives = 34/80 (42%), Gaps = 7/80 (8%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNA 119
R G G G G G G G GRG G G RG GRGG + G+ + +
Sbjct: 41 RGRGRGGGGGGGGGGG-----GGRGGGGFHSGGNRGRGRGG--KRGNQSGKNVMVEPHRH 93
Query: 120 DGLYVGDNADGEKLAKKLGP 139
+G+++ + + K L P
Sbjct: 94 EGVFICRGKEDALVTKNLVP 113
Score = 31.6 bits (70), Expect = 2.6
Identities = 25/75 (33%), Positives = 29/75 (38%), Gaps = 1/75 (1%)
Query: 60 RDDGDGSGRGRGRGRGRDVYA-RGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSN 118
R G G G G G G GR G+RGRG G RG+ G R I R
Sbjct: 43 RGRGGGGGGGGGGGGGRGGGGFHSGGNRGRGRGGKRGNQSGKNVMVEPHRHEGVFICRGK 102
Query: 119 ADGLYVGDNADGEKL 133
D L + GE +
Sbjct: 103 EDALVTKNLVPGESV 117
>DHX8_ARATH (Q38953) Putative pre-mRNA splicing factor ATP-dependent
RNA helicase
Length = 1168
Score = 42.0 bits (97), Expect = 0.002
Identities = 58/255 (22%), Positives = 97/255 (37%), Gaps = 24/255 (9%)
Query: 2 TVPKVLPGGGRGRGKPLEEAAQEAPQAPVVNRHIRVRQTPADAESDNVPRRQPMNRFVRD 61
T+ + P + K E Q+ + + +V++ + E + RR+ +R
Sbjct: 72 TIHGIYPPKPKSEKKKEEGDDQKFKGLAIKDTKDKVKELEKEIEREAEERRREEDRNRDR 131
Query: 62 DGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDRTSHQDIARSNADG 121
D SGR R R R RD R DR R RGD G D R+ + R DG
Sbjct: 132 DRRESGRDRDRDRNRD--RDDRRDRHRDRERNRGDEEG-----EDRRSDRRHRERGRGDG 184
Query: 122 LYVGDNADGEKLAKKLGPEIMDQITEAYEEIIERVLPSPLQDEYVEAMDINCAIEFEPEY 181
G+ D + + + + A E + +V + MD C ++F+
Sbjct: 185 ---GEGEDRRRDRRAKDEYVEEDKGGANEPELYQV----YKGRVTRVMDAGCFVQFDKFR 237
Query: 182 AVEFDNPDIDEKEPIALRDALEKMKPFLMTYEGIRSQEEWEEVIEELMQRVPLLKKIVDH 241
E + +A R ++K K F+ R E + +VI + L + VD
Sbjct: 238 GKE----GLVHVSQMATR-RVDKAKEFVK-----RDMEVYVKVISISSDKYSLSMRDVDQ 287
Query: 242 YSGPDRVTAKKQQEE 256
+G D + +K +E
Sbjct: 288 NTGRDLIPLRKPSDE 302
>RS2_CAEEL (P51403) 40S ribosomal protein S2
Length = 272
Score = 41.6 bits (96), Expect = 0.003
Identities = 23/42 (54%), Positives = 25/42 (58%), Gaps = 1/42 (2%)
Query: 67 GRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYGDDR 108
GRG GRG R R G GRGGRGGRG GRGG G ++
Sbjct: 13 GRGGGRGGARPAGDRPAGRGGRGGRGGRG-GRGGRAGRGGEK 53
Score = 38.9 bits (89), Expect = 0.016
Identities = 24/41 (58%), Positives = 24/41 (58%), Gaps = 3/41 (7%)
Query: 65 GSGRGRGRGRGRDVYARGRGDR-GRGGRGGRGD--GRGGFK 102
G G GRG R GRG R GRGGRGGRG GRGG K
Sbjct: 13 GRGGGRGGARPAGDRPAGRGGRGGRGGRGGRGGRAGRGGEK 53
>NOA1_HUMAN (Q9NY12) Nucleolar protein family A member 1 (snoRNP
protein GAR1) (H/ACA ribonucleoprotein GAR1)
Length = 217
Score = 41.6 bits (96), Expect = 0.003
Identities = 26/53 (49%), Positives = 29/53 (54%), Gaps = 4/53 (7%)
Query: 54 PMNRFVRDDGDGSGRGRGRGRGRDVYARGRGDR--GRGG--RGGRGDGRGGFK 102
P+ RF+ G RG GRG RG G R GRGG RGGRG G GGF+
Sbjct: 153 PLQRFLPRPPGEKGPPRGGGRGGRGGGRGGGGRGGGRGGGFRGGRGGGGGGFR 205
Score = 37.4 bits (85), Expect = 0.048
Identities = 22/41 (53%), Positives = 24/41 (57%), Gaps = 6/41 (14%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGRGGFKRYG 105
G GRG GRG G + GRG G G RGGRG GGF+ G
Sbjct: 182 GGGRGGGRGGG---FRGGRGGGGGGFRGGRG---GGFRGRG 216
Score = 33.5 bits (75), Expect = 0.69
Identities = 24/49 (48%), Positives = 25/49 (50%), Gaps = 8/49 (16%)
Query: 65 GSGRGR-GRGRGRDVYARGRGD---RGRGGRGG----RGDGRGGFKRYG 105
G GRG RG G + RG RG GG GG RG GRGGF R G
Sbjct: 5 GGGRGGFNRGGGGGGFNRGGSSNHFRGGGGGGGGGNFRGGGRGGFGRGG 53
Score = 32.3 bits (72), Expect = 1.5
Identities = 21/49 (42%), Positives = 24/49 (48%), Gaps = 7/49 (14%)
Query: 65 GSGRGRGRGRGRDVYARGRGDRGRG-----GRGG--RGDGRGGFKRYGD 106
G G G RG + + G G G G GRGG RG GRGGF + D
Sbjct: 15 GGGGGFNRGGSSNHFRGGGGGGGGGNFRGGGRGGFGRGGGRGGFNKGQD 63
Score = 30.0 bits (66), Expect = 7.6
Identities = 17/39 (43%), Positives = 19/39 (48%), Gaps = 6/39 (15%)
Query: 60 RDDGDGSGRGRGRGRGRDVYARGRGDRGRGGRGGRGDGR 98
R G G G G RG GR + RG GGRGG G+
Sbjct: 30 RGGGGGGGGGNFRGGGRGGFGRG------GGRGGFNKGQ 62
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.137 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 41,864,357
Number of Sequences: 164201
Number of extensions: 2174319
Number of successful extensions: 15622
Number of sequences better than 10.0: 382
Number of HSP's better than 10.0 without gapping: 193
Number of HSP's successfully gapped in prelim test: 205
Number of HSP's that attempted gapping in prelim test: 8306
Number of HSP's gapped (non-prelim): 4040
length of query: 310
length of database: 59,974,054
effective HSP length: 110
effective length of query: 200
effective length of database: 41,911,944
effective search space: 8382388800
effective search space used: 8382388800
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147483.4


