

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147482.11 - phase: 0
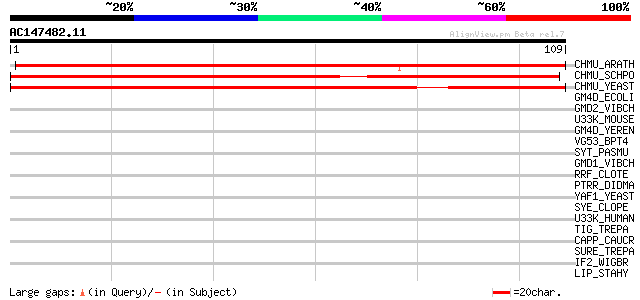
(109 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CHMU_ARATH (P42738) Chorismate mutase, chloroplast precursor (EC... 152 1e-37
CHMU_SCHPO (O13739) Probable chorismate mutase (EC 5.4.99.5) (CM) 89 2e-18
CHMU_YEAST (P32178) Chorismate mutase (EC 5.4.99.5) (CM) 81 4e-16
GM4D_ECOLI (P32054) GDP-mannose 4,6-dehydratase (EC 4.2.1.47) (G... 33 0.12
GMD2_VIBCH (Q56598) Probable GDP-mannose 4,6-dehydratase (EC 4.2... 32 0.35
U33K_MOUSE (Q922Y1) UBA/UBX 33.3 kDa protein 31 0.59
GM4D_YEREN (Q56872) Probable GDP-mannose 4,6-dehydratase (EC 4.2... 30 0.77
VG53_BPT4 (P16011) Baseplate structural protein Gp53 (Baseplate ... 30 1.0
SYT_PASMU (P57857) Threonyl-tRNA synthetase (EC 6.1.1.3) (Threon... 30 1.0
GMD1_VIBCH (Q06952) Probable GDP-mannose 4,6-dehydratase (EC 4.2... 30 1.0
RRF_CLOTE (Q895K9) Ribosome recycling factor (Ribosome releasing... 30 1.3
PTRR_DIDMA (P25107) Parathyroid hormone/parathyroid hormone-rela... 30 1.3
YAF1_YEAST (P39720) Putative 118.2 kDa transcriptional regulator... 29 1.7
SYE_CLOPE (Q8XLP3) Glutamyl-tRNA synthetase (EC 6.1.1.17) (Gluta... 29 1.7
U33K_HUMAN (Q04323) UBA/UBX 33.3 kDa protein 29 2.2
TIG_TREPA (O83519) Trigger factor (TF) 29 2.2
CAPP_CAUCR (Q9A871) Phosphoenolpyruvate carboxylase (EC 4.1.1.31... 29 2.2
SURE_TREPA (O83434) Acid phosphatase surE (EC 3.1.3.2) 28 2.9
IF2_WIGBR (Q8D2X6) Translation initiation factor IF-2 28 2.9
LIP_STAHY (P04635) Lipase precursor (EC 3.1.1.3) (Triacylglycero... 28 3.8
>CHMU_ARATH (P42738) Chorismate mutase, chloroplast precursor (EC
5.4.99.5) (CM-1)
Length = 334
Score = 152 bits (385), Expect = 1e-37
Identities = 75/115 (65%), Positives = 95/115 (82%), Gaps = 7/115 (6%)
Query: 2 LSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYG 61
LSKRIHYGKFVAEAKFQA+P++Y++AI AQDKD LM++LT+P VE++IK+RV +K +TYG
Sbjct: 220 LSKRIHYGKFVAEAKFQASPEAYESAIKAQDKDALMDMLTFPTVEDAIKKRVEMKTRTYG 279
Query: 62 QEVAVNLKDQKTEP-------VYKINPSLVADLYSDWIMPLTKEVQVAYLLRKLD 109
QEV V +++++ E VYKI+P LV DLY DWIMPLTKEVQV YLLR+LD
Sbjct: 280 QEVKVGMEEKEEEEEEGNESHVYKISPILVGDLYGDWIMPLTKEVQVEYLLRRLD 334
>CHMU_SCHPO (O13739) Probable chorismate mutase (EC 5.4.99.5) (CM)
Length = 251
Score = 89.0 bits (219), Expect = 2e-18
Identities = 46/108 (42%), Positives = 69/108 (63%), Gaps = 5/108 (4%)
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
+LS+RIHYGKFVAEAK+ A P+ YK I+A+D + + EE + +R+ KA Y
Sbjct: 148 SLSRRIHYGKFVAEAKYLANPEKYKKLILARDIKGIENEIVDAAQEERVLKRLHYKALNY 207
Query: 61 GQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEVQVAYLLRKL 108
G++ A T+P +IN VA +Y D+++P+TK+V+V YLL +L
Sbjct: 208 GRDAA-----DPTKPSDRINADCVASIYKDYVIPMTKKVEVDYLLARL 250
>CHMU_YEAST (P32178) Chorismate mutase (EC 5.4.99.5) (CM)
Length = 256
Score = 81.3 bits (199), Expect = 4e-16
Identities = 43/109 (39%), Positives = 68/109 (61%), Gaps = 6/109 (5%)
Query: 1 ALSKRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
+LS+RIH+GKFVAEAKFQ+ Y I ++D + +M+ +T VEE I R+ KA+ Y
Sbjct: 153 SLSRRIHFGKFVAEAKFQSDIPLYTKLIKSKDVEGIMKNITNSAVEEKILERLTKKAEVY 212
Query: 61 GQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEVQVAYLLRKLD 109
G + +++ P Y + +Y + ++P+TKEV+V YLLR+L+
Sbjct: 213 GVDPTNESGERRITPEYLVK------IYKEIVIPITKEVEVEYLLRRLE 255
>GM4D_ECOLI (P32054) GDP-mannose 4,6-dehydratase (EC 4.2.1.47)
(GDP-D-mannose dehydratase)
Length = 373
Score = 33.1 bits (74), Expect = 0.12
Identities = 20/69 (28%), Positives = 35/69 (49%), Gaps = 2/69 (2%)
Query: 24 YKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSL 83
Y A + A RL+E + + +E+ + A ++ YG + + ++T P Y +P
Sbjct: 101 YTADVDAMGTLRLLEAIRFLGLEKKTRFYQASTSELYG--LVQEIPQKETTPFYPRSPYA 158
Query: 84 VADLYSDWI 92
VA LY+ WI
Sbjct: 159 VAKLYAYWI 167
>GMD2_VIBCH (Q56598) Probable GDP-mannose 4,6-dehydratase (EC
4.2.1.47) (GDP-D-mannose dehydratase)
Length = 372
Score = 31.6 bits (70), Expect = 0.35
Identities = 20/70 (28%), Positives = 35/70 (49%), Gaps = 2/70 (2%)
Query: 24 YKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSL 83
Y A + A RL+E + +E++ K A ++ YG + ++T P Y +P
Sbjct: 101 YTADVDAMGTLRLLEAIRLLGLEKTTKFYQASTSELYG--LVQETPQKETTPFYPRSPYA 158
Query: 84 VADLYSDWIM 93
VA +Y+ WI+
Sbjct: 159 VAKMYAYWIV 168
>U33K_MOUSE (Q922Y1) UBA/UBX 33.3 kDa protein
Length = 297
Score = 30.8 bits (68), Expect = 0.59
Identities = 19/72 (26%), Positives = 33/72 (45%)
Query: 4 KRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQE 63
+R G+ ++ A+ + D + A + +++ EL V E I+R A +AK YG
Sbjct: 123 QRRRQGQELSVARQKLQEDEMRRAAEERRREKAEELAARQRVREKIERDKAERAKKYGGS 182
Query: 64 VAVNLKDQKTEP 75
V T+P
Sbjct: 183 VGSRSSPPATDP 194
>GM4D_YEREN (Q56872) Probable GDP-mannose 4,6-dehydratase (EC
4.2.1.47) (GDP-D-mannose dehydratase) (ORF13.7)
Length = 372
Score = 30.4 bits (67), Expect = 0.77
Identities = 19/69 (27%), Positives = 33/69 (47%), Gaps = 2/69 (2%)
Query: 24 YKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSL 83
Y A + A RL+E + +E + A ++ YG + + ++T P Y +P
Sbjct: 100 YTADVDAMGTLRLLEAVRINGLEHKTRFYQASTSELYG--LVQEIPQRETTPFYPRSPYA 157
Query: 84 VADLYSDWI 92
VA +Y+ WI
Sbjct: 158 VAKMYAYWI 166
>VG53_BPT4 (P16011) Baseplate structural protein Gp53 (Baseplate
wedge protein 53)
Length = 196
Score = 30.0 bits (66), Expect = 1.0
Identities = 11/45 (24%), Positives = 27/45 (59%)
Query: 19 AAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQE 63
AA D+Y+AA++ +K R ++++ ++ + + + K+YG +
Sbjct: 151 AAVDTYEAAVLENEKLRQIKIIAKSDINSFMNDLIRIMEKSYGND 195
>SYT_PASMU (P57857) Threonyl-tRNA synthetase (EC 6.1.1.3)
(Threonine--tRNA ligase) (ThrRS)
Length = 643
Score = 30.0 bits (66), Expect = 1.0
Identities = 17/75 (22%), Positives = 36/75 (47%), Gaps = 10/75 (13%)
Query: 19 AAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYG----------QEVAVNL 68
A P A+ +A+D DR ++ + + SI+R + + + Y Q + +N+
Sbjct: 488 ALPGRLNASYVAEDNDRRTPVMIHRAILGSIERFIGIITEEYAGFFPAWLAPTQAIVMNI 547
Query: 69 KDQKTEPVYKINPSL 83
D +++ V K+ +L
Sbjct: 548 TDSQSDYVQKVVKTL 562
>GMD1_VIBCH (Q06952) Probable GDP-mannose 4,6-dehydratase (EC
4.2.1.47) (GDP-D-mannose dehydratase)
Length = 373
Score = 30.0 bits (66), Expect = 1.0
Identities = 19/69 (27%), Positives = 34/69 (48%), Gaps = 2/69 (2%)
Query: 24 YKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSL 83
Y A + A RL+E + + + + K A ++ YG + + ++T P Y +P
Sbjct: 102 YTADVDAIGTLRLLEAIRFLGLTKKTKFYQASTSELYG--LVQEIPQKETTPFYPRSPYA 159
Query: 84 VADLYSDWI 92
VA +Y+ WI
Sbjct: 160 VAKMYAYWI 168
>RRF_CLOTE (Q895K9) Ribosome recycling factor (Ribosome releasing
factor) (RRF)
Length = 183
Score = 29.6 bits (65), Expect = 1.3
Identities = 16/61 (26%), Positives = 30/61 (48%), Gaps = 1/61 (1%)
Query: 40 LTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEV 99
L PE+ E +R + K YG++ + ++ + + KI L +D+ D I +E+
Sbjct: 100 LIIPELTEETRRNIVKTVKKYGEDTKIAIRSVRRDGNDKIK-DLKSDMSEDDIKKAEEEI 158
Query: 100 Q 100
Q
Sbjct: 159 Q 159
>PTRR_DIDMA (P25107) Parathyroid hormone/parathyroid
hormone-related peptide receptor precursor (PTH/PTHr
receptor) (PTH/PTHrP type I receptor)
Length = 585
Score = 29.6 bits (65), Expect = 1.3
Identities = 18/45 (40%), Positives = 25/45 (55%)
Query: 30 AQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTE 74
AQ + RL E+L PE+ ES K ++ AKT ++ A L Q E
Sbjct: 46 AQCEQRLKEVLRVPELAESAKDWMSRSAKTKKEKPAEKLYSQAEE 90
>YAF1_YEAST (P39720) Putative 118.2 kDa transcriptional regulatory
protein in ACS1-GCV3 intergenic region
Length = 1062
Score = 29.3 bits (64), Expect = 1.7
Identities = 17/47 (36%), Positives = 24/47 (50%), Gaps = 1/47 (2%)
Query: 33 KDRLMELLTYPEVEESIKRRVA-VKAKTYGQEVAVNLKDQKTEPVYK 78
KD+ M+LLT EV ES KR + + + K K EP+Y+
Sbjct: 123 KDKAMKLLTEREVNESGKRSASPINTNNASGDSPDTKKQHKMEPIYE 169
>SYE_CLOPE (Q8XLP3) Glutamyl-tRNA synthetase (EC 6.1.1.17)
(Glutamate--tRNA ligase) (GluRS)
Length = 546
Score = 29.3 bits (64), Expect = 1.7
Identities = 12/34 (35%), Positives = 19/34 (55%)
Query: 66 VNLKDQKTEPVYKINPSLVADLYSDWIMPLTKEV 99
V L D + + K+ P +V DLY++W KE+
Sbjct: 361 VKLNDVSKDVICKMKPEVVYDLYTNWAKEYDKEM 394
>U33K_HUMAN (Q04323) UBA/UBX 33.3 kDa protein
Length = 297
Score = 28.9 bits (63), Expect = 2.2
Identities = 19/72 (26%), Positives = 32/72 (44%)
Query: 4 KRIHYGKFVAEAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTYGQE 63
+R G+ ++ A+ + D + A + +++ EL V E I+R A +AK YG
Sbjct: 123 QRRRQGQELSAARQRLQEDEMRRAAEERRREKAEELAARQRVREKIERDKAERAKKYGGS 182
Query: 64 VAVNLKDQKTEP 75
V EP
Sbjct: 183 VGSQPPPVAPEP 194
>TIG_TREPA (O83519) Trigger factor (TF)
Length = 442
Score = 28.9 bits (63), Expect = 2.2
Identities = 14/34 (41%), Positives = 24/34 (70%)
Query: 43 PEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPV 76
PEVEE +K+RV V+ ++V+V+ ++ +TE V
Sbjct: 346 PEVEEHLKQRVIVELLLKQEQVSVSAEEIETEYV 379
>CAPP_CAUCR (Q9A871) Phosphoenolpyruvate carboxylase (EC 4.1.1.31)
(PEPCase) (PEPC)
Length = 909
Score = 28.9 bits (63), Expect = 2.2
Identities = 20/68 (29%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query: 44 EVEESIKRRVAVKAKT--YGQEVAVNLKDQKTEPVYKINPSL---VADLYSDWIMPLTKE 98
E+ ES+ R +A+ +T Y E + +KD+ + + S+ + DLY DW + +
Sbjct: 185 EIRESLFREIALMWRTRLYRPE-RITVKDEIRNALSIVRTSILPAIIDLYGDWTGKIGQH 243
Query: 99 VQVAYLLR 106
Q+A LL+
Sbjct: 244 GQLAPLLK 251
>SURE_TREPA (O83434) Acid phosphatase surE (EC 3.1.3.2)
Length = 187
Score = 28.5 bits (62), Expect = 2.9
Identities = 15/43 (34%), Positives = 23/43 (52%), Gaps = 8/43 (18%)
Query: 15 AKFQAAPDSYKAAIIAQDKDR--------LMELLTYPEVEESI 49
A +AAP+ Y+ ++A D+DR +E +T EVE I
Sbjct: 21 AALKAAPEGYEVTVVAPDRDRSAVSHGITTLEPVTVKEVEPGI 63
>IF2_WIGBR (Q8D2X6) Translation initiation factor IF-2
Length = 841
Score = 28.5 bits (62), Expect = 2.9
Identities = 23/102 (22%), Positives = 47/102 (45%), Gaps = 5/102 (4%)
Query: 5 RIHYGKFVA----EAKFQAAPDSYKAAIIAQDKDRLMELLTYPEVEESIKRRVAVKAKTY 60
R+H G V K +A DS+K I + +E+L + S + VK +
Sbjct: 542 RLHQGDVVLCGCEYGKIKAMQDSFKCKIKSVGPSIPVEILGLSGIPISGDKITVVKEEKK 601
Query: 61 GQEVAVNLKDQKTEPVYKINPSL-VADLYSDWIMPLTKEVQV 101
+EVA+ +++ E +IN + + +++S+ + K + +
Sbjct: 602 AREVAIYRQNKLRESKLRINKKIKIENVFSNLNLSTKKNINI 643
>LIP_STAHY (P04635) Lipase precursor (EC 3.1.1.3) (Triacylglycerol
lipase)
Length = 641
Score = 28.1 bits (61), Expect = 3.8
Identities = 16/45 (35%), Positives = 21/45 (46%)
Query: 34 DRLMELLTYPEVEESIKRRVAVKAKTYGQEVAVNLKDQKTEPVYK 78
D E PE ES K V V+ ++ QEV K + +P YK
Sbjct: 78 DDSKEATPLPEKAESPKTEVTVQPSSHTQEVPALHKKTQQQPAYK 122
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,415,877
Number of Sequences: 164201
Number of extensions: 383674
Number of successful extensions: 1211
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 1187
Number of HSP's gapped (non-prelim): 41
length of query: 109
length of database: 59,974,054
effective HSP length: 85
effective length of query: 24
effective length of database: 46,016,969
effective search space: 1104407256
effective search space used: 1104407256
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC147482.11


