

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.4 + phase: 0
(274 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
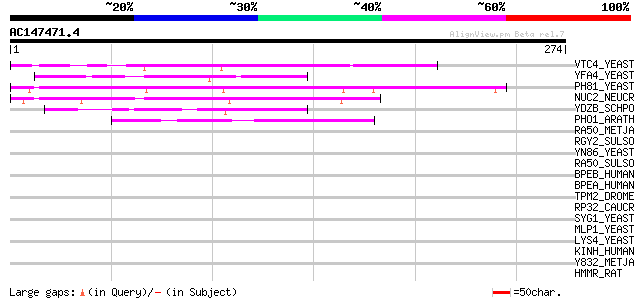
Score E
Sequences producing significant alignments: (bits) Value
VTC4_YEAST (P47075) VTC4 protein 71 2e-12
YFA4_YEAST (P43585) Hypothetical 95.4 kDa protein in SEC4-MSH4 i... 59 1e-08
PH81_YEAST (P17442) Phosphate system positive regulatory protein... 58 2e-08
NUC2_NEUCR (Q01317) Ankyrin repeat protein nuc-2 56 1e-07
YDZB_SCHPO (O13718) Hypothetical protein C14C4.11 in chromosome I 47 4e-05
PHO1_ARATH (Q8S403) Putative phosphate transporter 1 44 6e-04
RA50_METJA (Q58718) DNA double-strand break repair rad50 ATPase 39 0.014
RGY2_SULSO (Q97ZF5) Reverse gyrase 2 [Includes: Helicase (EC 3.6... 36 0.12
YN86_YEAST (P27514) Hypothetical 99.5 kDa protein in URK1-SMM1 i... 35 0.26
RA50_SULSO (Q97WH0) DNA double-strand break repair rad50 ATPase 35 0.26
BPEB_HUMAN (Q8WXK8) Bullous pemphigoid antigen 1, isoform 7 (Bul... 34 0.34
BPEA_HUMAN (O94833) Bullous pemphigoid antigen 1, isoforms 6/9/1... 34 0.34
TPM2_DROME (P09491) Tropomyosin 2 (Tropomyosin I) 34 0.44
RP32_CAUCR (P48194) RNA polymerase sigma-32 factor 33 0.75
SYG1_YEAST (P40528) SYG1 protein 33 0.98
MLP1_YEAST (Q02455) MLP1 protein (Myosin-like protein 1) 33 0.98
LYS4_YEAST (P49367) Homoaconitase, mitochondrial precursor (EC 4... 33 0.98
KINH_HUMAN (P33176) Kinesin heavy chain (Ubiquitous kinesin heav... 32 1.3
Y832_METJA (Q58242) Hypothetical protein MJ0832 [Contains: Mja r... 32 1.7
HMMR_RAT (P97779) Hyaluronan mediated motility receptor (Intrace... 32 1.7
>VTC4_YEAST (P47075) VTC4 protein
Length = 721
Score = 71.2 bits (173), Expect = 2e-12
Identities = 61/220 (27%), Positives = 105/220 (47%), Gaps = 30/220 (13%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEV 60
MKF + L+ + + + ++SY DLK E++ + SK G+
Sbjct: 1 MKFGEHLSKSL---IRQYSYYYISYDDLKT--------ELEDNLSKNN---------GQW 40
Query: 61 TKEVK-DFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWA--------KSSDIELM 111
T+E++ DFL LE+E++K F K E + KE+Q++V + ++
Sbjct: 41 TQELETDFLESLEIELDKVYTFCKVKHSEVFRRVKEVQEQVQHTVRLLDSNNPPTQLDFE 100
Query: 112 TVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKIDV 171
+ E+ D ++ L +S LNYTG KIIKK+DK+TG +L+ F + ++PFFK +
Sbjct: 101 ILEEELSDIIADVHDLAKFSRLNYTGFQKIIKKHDKKTGFILKPVFQVRLDSKPFFK-EN 159
Query: 172 LNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETT 211
++LV + + I P+ S + R+TT
Sbjct: 160 YDELVVKISQLYDIARTSGRPIKGDSSAGGKQQNFVRQTT 199
>YFA4_YEAST (P43585) Hypothetical 95.4 kDa protein in SEC4-MSH4
intergenic region
Length = 828
Score = 58.9 bits (141), Expect = 1e-08
Identities = 42/138 (30%), Positives = 68/138 (48%), Gaps = 16/138 (11%)
Query: 13 QTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLLE 72
+ P W+ +++Y+ LKK LK K D S K+ R DD ++ F+ L+
Sbjct: 10 EVYPPWKGSYINYEGLKKFLKEDSVK--DGSNDKKARWDDSDESK---------FVEELD 58
Query: 73 VEIEKFNGFFVEKEEEYVIKWKELQ---DKVAWAKSSDIELMTVGREIVDFHGEMVLLEN 129
E+EK GF ++K + + L+ D A K+ D + R + + E L+N
Sbjct: 59 KELEKVYGFQLKKYNNLMERLSHLEKQTDTEAAIKALDADAFQ--RVLEELLSESTELDN 116
Query: 130 YSALNYTGLVKIIKKYDK 147
+ LN+TG KI+KK+DK
Sbjct: 117 FKRLNFTGFAKIVKKHDK 134
>PH81_YEAST (P17442) Phosphate system positive regulatory protein
PHO81 (CDK inhibitor PHO81)
Length = 1178
Score = 58.2 bits (139), Expect = 2e-08
Identities = 66/263 (25%), Positives = 114/263 (43%), Gaps = 20/263 (7%)
Query: 1 MKFWKILN-NQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGE 59
MKF K L Q+E L ++ F+ YK LKK +K + + +S L D E
Sbjct: 1 MKFGKYLEARQLE--LAEYNSHFIDYKALKKLIKQLAIPTLKASSDLDLHLTLDDIDEKI 58
Query: 60 VTKEVKD----FLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAK-------SSDI 108
+ + +++ F LE E+EK NG+++ +E + IK+ L K K +
Sbjct: 59 IHQRLQENKAAFFFKLERELEKVNGYYLARESDLRIKFNILHSKYKDYKINGKLNSNQAT 118
Query: 109 ELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLN-QPFF 167
+ F ++ LE Y LN TG K +KK+DKR+ + + ++ V++ QP F
Sbjct: 119 SFKNLYAAFKKFQKDLRNLEQYVELNKTGFSKALKKWDKRSQSHDKDFYLATVVSIQPIF 178
Query: 168 KIDVLNKLVKE-CEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKE 226
D KL E ++L + + + +S + + T+N K ++
Sbjct: 179 TRDGPLKLNDETLHILLELNDIDNNNRRADIQSSTFTNDDDDDNNTSNNNKHNNNNNNNN 238
Query: 227 FSEIQNMENIFI----KLTTSAL 245
+ N N + +LTTS +
Sbjct: 239 NNNNNNNNNNILHNNYELTTSKI 261
>NUC2_NEUCR (Q01317) Ankyrin repeat protein nuc-2
Length = 1066
Score = 55.8 bits (133), Expect = 1e-07
Identities = 51/197 (25%), Positives = 89/197 (44%), Gaps = 20/197 (10%)
Query: 1 MKFWK-ILNNQIEQTLPDWRDKFLSYKDLKKQLKL-----IVPKEIDSSCSKRRRLDDDG 54
MKF K I Q+E +P++ F++YK LKK +K I+P + D + LD
Sbjct: 1 MKFGKQIQKRQLE--VPEYAASFVNYKALKKLIKKLSATPILPPQTDLRRAPGEPLDTQS 58
Query: 55 GAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSD------- 107
+ F ++ E++K N +V+KE E I+ K L DK +S
Sbjct: 59 ALQANKAT----FFFQIDRELDKVNACYVQKEAELKIRLKTLLDKKKALRSRSGGTSRRS 114
Query: 108 IELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVL-NQPF 166
+ T+ F ++ L+ + +N T KI+KK+DK + + ++ V+ +P
Sbjct: 115 TKFTTLQEGFQQFVNDLNKLQQFVEINGTAFSKILKKWDKTAKSKTKELYLSRVVEKRPA 174
Query: 167 FKIDVLNKLVKECEVML 183
F V+++L + L
Sbjct: 175 FNPTVISELSDQATTSL 191
>YDZB_SCHPO (O13718) Hypothetical protein C14C4.11 in chromosome I
Length = 734
Score = 47.4 bits (111), Expect = 4e-05
Identities = 38/143 (26%), Positives = 62/143 (42%), Gaps = 38/143 (26%)
Query: 18 WRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLLEVEIEK 77
WRDK+++Y +LK LK +++ + GE + F+ +++ ++EK
Sbjct: 15 WRDKYMNYPELKHLLKT----------------EEEAPSWGE--NDESKFVSVMDAQLEK 56
Query: 78 FNGFFVEKEEEYVIKWKELQDKVAWAKS-------------SDIELMTVGREIVDFHGEM 124
F +E KEL + V W KS S E + + + +
Sbjct: 57 VYAFHLEI-------LKELNESVDWVKSKVSASQEPDGPPISKEEAIKLLERLDSCTETV 109
Query: 125 VLLENYSALNYTGLVKIIKKYDK 147
LE Y+ LN TG KI+KK+DK
Sbjct: 110 KKLEKYTRLNLTGFFKIVKKHDK 132
>PHO1_ARATH (Q8S403) Putative phosphate transporter 1
Length = 782
Score = 43.5 bits (101), Expect = 6e-04
Identities = 33/130 (25%), Positives = 55/130 (41%), Gaps = 17/130 (13%)
Query: 51 DDDGGAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIEL 110
D GAEG + + + + +L E+ V WK +Q +S+ +EL
Sbjct: 243 DAVAGAEGGIARSIATAMSVLWEEL-------VNNPRSDFTNWKNIQSAEKKIRSAFVEL 295
Query: 111 MTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKID 170
+ + LL+ YS+LN KI+KK+DK G +++ V F D
Sbjct: 296 ----------YRGLGLLKTYSSLNMIAFTKIMKKFDKVAGQNASSTYLKVVKRSQFISSD 345
Query: 171 VLNKLVKECE 180
+ +L+ E E
Sbjct: 346 KVVRLMDEVE 355
Score = 30.4 bits (67), Expect = 4.8
Identities = 15/35 (42%), Positives = 24/35 (67%), Gaps = 3/35 (8%)
Query: 1 MKFWKILNNQIEQTLPDWRDKFLSYKDLKKQLKLI 35
+KF K L Q+ +P+W++ F++Y LKKQ+K I
Sbjct: 2 VKFSKELEAQL---IPEWKEAFVNYCLLKKQIKKI 33
>RA50_METJA (Q58718) DNA double-strand break repair rad50 ATPase
Length = 1005
Score = 38.9 bits (89), Expect = 0.014
Identities = 55/249 (22%), Positives = 106/249 (42%), Gaps = 30/249 (12%)
Query: 10 QIEQTLPDWRDKFLSYKDLKKQLKLIVP-----KEIDSSCSKRRRLDD---------DGG 55
+IE L + + + Y L KQL++I KE + R +D+ D
Sbjct: 298 KIESRLRELKSHYEDYLKLTKQLEIIKGDIEKLKEFINKSKYRDDIDNLDTLLNKIKDEI 357
Query: 56 AEGEVTKEVKDFLRLLEVEIEKFNGF--FVEKEEEYVIKWKELQDKVAWAKSSDIELMT- 112
E K++ + L+ L EIEK + E+ +EY K+ EL++K +E +T
Sbjct: 358 ERVETIKDLLEELKNLNEEIEKIEKYKRICEECKEYYEKYLELEEKAVEYNKLTLEYITL 417
Query: 113 ------VGREIVDFHGEM-VLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQP 165
+ + I D + LLE ++ + +K+ +++ L L + LN+
Sbjct: 418 LQEKKSIEKNINDLETRINKLLEETKNIDIESIENSLKEIEEKKKVLENLQKEKIELNK- 476
Query: 166 FFKIDVLNKLVKECEVMLSIIFPKSG--PLGQS-LSTSEVFEEVARETTTANETKETLDH 222
K+ +N +K + +L + G PL ++ + ++ E + + T N L+
Sbjct: 477 --KLGEINSEIKRLKKILDELKEVEGKCPLCKTPIDENKKMELINQHKTQLNNKYTELEE 534
Query: 223 VPKEFSEIQ 231
+ K+ EI+
Sbjct: 535 INKKIREIE 543
>RGY2_SULSO (Q97ZF5) Reverse gyrase 2 [Includes: Helicase (EC
3.6.1.-); Topoisomerase (EC 5.99.1.3)]
Length = 1166
Score = 35.8 bits (81), Expect = 0.12
Identities = 32/121 (26%), Positives = 56/121 (45%), Gaps = 15/121 (12%)
Query: 36 VPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKEEEYVIKWKE 95
+PK++ S + RL + E T+EVK+ +R LE++I +F +K + VI
Sbjct: 211 IPKDVYESAIQLIRLRNKYYFSNEYTEEVKEKIRELELKIAEFK----DKISQLVIASAT 266
Query: 96 LQDKVAWAKSSDIELMT---------VGREIVDFHGEMVLLENYSALNYTGLVKIIKKYD 146
++ K K + L+T R I+D + E + L L GL+ + K+Y
Sbjct: 267 IRPK--GIKQKALRLLTGFEPSSIQLYARNIIDAYTENLDLSIVKELGSGGLILVSKEYG 324
Query: 147 K 147
+
Sbjct: 325 R 325
>YN86_YEAST (P27514) Hypothetical 99.5 kDa protein in URK1-SMM1
intergenic region
Length = 894
Score = 34.7 bits (78), Expect = 0.26
Identities = 21/96 (21%), Positives = 43/96 (43%), Gaps = 9/96 (9%)
Query: 14 TLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEG---------EVTKEV 64
++P+W K+L+Y LKK + + ++ S+ D E + +
Sbjct: 11 SVPEWSTKYLAYSQLKKLIYSLQKDKLYSNNKHHVVEPHDANDENLPLLADASPDDQFYI 70
Query: 65 KDFLRLLEVEIEKFNGFFVEKEEEYVIKWKELQDKV 100
F+ L E++K + F++ +E + + EL+D V
Sbjct: 71 SKFVAALNQELKKIDKFYISQETGLIANYNELKDDV 106
Score = 31.6 bits (70), Expect = 2.2
Identities = 18/63 (28%), Positives = 33/63 (51%), Gaps = 4/63 (6%)
Query: 111 MTVGREIVDFHGEMVLLENYSALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKID 170
+++ + ++ + ++ L+++ LN TG KI KK+DK L+ Q+ LN F
Sbjct: 208 LSLKKRLISIYTQLSELKDFIELNQTGFSKICKKFDKSLNTNLK----QNYLNYIKFHSH 263
Query: 171 VLN 173
V N
Sbjct: 264 VFN 266
>RA50_SULSO (Q97WH0) DNA double-strand break repair rad50 ATPase
Length = 864
Score = 34.7 bits (78), Expect = 0.26
Identities = 53/242 (21%), Positives = 100/242 (40%), Gaps = 29/242 (11%)
Query: 21 KFLSYKDLKKQLKLIVPK---------EIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLL 71
K+L YK+L+++L+ + PK ++DS + + RL+ D L
Sbjct: 328 KYLKYKELERKLEELQPKYQQYLKLKSDLDSKLNLKERLEKDASE--------------L 373
Query: 72 EVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDFHGEMVLLENYS 131
+I+K N +K EE K L+ ++A +S E + I GE +
Sbjct: 374 SNDIDKVNS-LEQKVEETRKKQLNLRAQLAKVESLISEKNEIINNISQVEGETCPVCG-R 431
Query: 132 ALNYTGLVKIIKKYDKRTGALLRLPFIQDVLNQPFFKI-DVLNKLVKECEVMLSIIFPKS 190
L+ KIIK + +L+L ++ L + KI + LNK+ +E + +
Sbjct: 432 PLDEEHKQKIIK---EAKSYILQLELNKNELEEELKKITNELNKIEREYRRLSNNKASYD 488
Query: 191 GPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFSEIQNMENIFIKLTTSALDTLKE 250
+ Q +E E + E + E + + +E E++ F++L+ + L +
Sbjct: 489 NVMRQLKKLNEEIENLHSEIESLKNIDEEIKKINEEVKELKLYYEEFMRLSKYTKEELDK 548
Query: 251 IR 252
R
Sbjct: 549 KR 550
>BPEB_HUMAN (Q8WXK8) Bullous pemphigoid antigen 1, isoform 7
(Bullous pemphigoid antigen) (BPA) (Hemidesmosomal
plaque protein) (Dystonia musculorum protein) (Dystonin)
(Fragment)
Length = 3060
Score = 34.3 bits (77), Expect = 0.34
Identities = 30/123 (24%), Positives = 54/123 (43%), Gaps = 9/123 (7%)
Query: 153 LRLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTT 212
+R P +D L++ F+I KL KE E + LG + E F A +++
Sbjct: 764 IRTPLERDDLHESVFRITEQEKLKKELERL-------KDDLGTITNKCEEFFSQAAASSS 816
Query: 213 ANETKETLDHVPKEFSEIQNMENIFI-KLTTSALDTLKEIRGGSSTVSIYSLPPLHSETL 271
+ L+ V + +++ +M + +I KL T L LK + + V +Y E +
Sbjct: 817 VPTLRSELNVVLQNMNQVYSMSSTYIDKLKTVNLG-LKNTQAAEALVKLYETKLCEEEAV 875
Query: 272 VED 274
+ D
Sbjct: 876 IAD 878
>BPEA_HUMAN (O94833) Bullous pemphigoid antigen 1, isoforms 6/9/10
(Trabeculin-beta) (Bullous pemphigoid antigen) (BPA)
(Hemidesmosomal plaque protein) (Dystonia musculorum
protein) (Dystonin)
Length = 5171
Score = 34.3 bits (77), Expect = 0.34
Identities = 30/123 (24%), Positives = 54/123 (43%), Gaps = 9/123 (7%)
Query: 153 LRLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTT 212
+R P +D L++ F+I KL KE E + LG + E F A +++
Sbjct: 764 IRTPLERDDLHESVFRITEQEKLKKELERL-------KDDLGTITNKCEEFFSQAAASSS 816
Query: 213 ANETKETLDHVPKEFSEIQNMENIFI-KLTTSALDTLKEIRGGSSTVSIYSLPPLHSETL 271
+ L+ V + +++ +M + +I KL T L LK + + V +Y E +
Sbjct: 817 VPTLRSELNVVLQNMNQVYSMSSTYIDKLKTVNLG-LKNTQAAEALVKLYETKLCEEEAV 875
Query: 272 VED 274
+ D
Sbjct: 876 IAD 878
>TPM2_DROME (P09491) Tropomyosin 2 (Tropomyosin I)
Length = 284
Score = 33.9 bits (76), Expect = 0.44
Identities = 38/193 (19%), Positives = 81/193 (41%), Gaps = 9/193 (4%)
Query: 26 KDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDF-LRLLEVEIEKFNGFFVE 84
K + +KL ID + + + D ++ +EV+D + ++VEI+ V
Sbjct: 5 KKKMQAMKLEKDNAIDKADTCENQAKDANSRADKLNEEVRDLEKKFVQVEID-----LVT 59
Query: 85 KEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDFHGEMVLLENYSALNYTGLVKIIKK 144
+E+ EL++K +++ E+ T R++ ++ E S L++ +
Sbjct: 60 AKEQLEKANTELEEKEKLLTATESEVATQNRKVQQIEEDLEKSEERSTTAQQKLLEATQS 119
Query: 145 YDKRTGALLRLPFIQDVLNQPFFKIDVLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFE 204
D+ +++ Q ++D L +KE ++ KS + + L+ E
Sbjct: 120 ADENNRMC---KVLENRSQQDEERMDQLTNQLKEARMLAEDADTKSDEVSRKLAFVEDEL 176
Query: 205 EVARETTTANETK 217
EVA + + E+K
Sbjct: 177 EVAEDRVRSGESK 189
>RP32_CAUCR (P48194) RNA polymerase sigma-32 factor
Length = 295
Score = 33.1 bits (74), Expect = 0.75
Identities = 22/79 (27%), Positives = 39/79 (48%), Gaps = 4/79 (5%)
Query: 74 EIEKFNGFFVEKEEEYVI--KWKELQDKVAWAKSSDIELMTVGREIVDFHGEMVLLENYS 131
EI KF + K+EE+++ +WKE QD A K L V + + + G + +
Sbjct: 21 EIRKFP--MLSKDEEFMLAQRWKEHQDPQAAHKMVTSHLRLVAKIAMGYRGYGLPIGEVI 78
Query: 132 ALNYTGLVKIIKKYDKRTG 150
+ GL++ +KK++ G
Sbjct: 79 SEGNVGLMQAVKKFEPEKG 97
>SYG1_YEAST (P40528) SYG1 protein
Length = 902
Score = 32.7 bits (73), Expect = 0.98
Identities = 12/29 (41%), Positives = 20/29 (68%)
Query: 5 KILNNQIEQTLPDWRDKFLSYKDLKKQLK 33
K ++ E +P+WRDK++ YK KK+L+
Sbjct: 2 KFADHLTESAIPEWRDKYIDYKVGKKKLR 30
>MLP1_YEAST (Q02455) MLP1 protein (Myosin-like protein 1)
Length = 1875
Score = 32.7 bits (73), Expect = 0.98
Identities = 50/218 (22%), Positives = 99/218 (44%), Gaps = 26/218 (11%)
Query: 26 KDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLLEVEI-----EKFNG 80
K L+ Q +L P I+ K++ + E EV++++++ L+ I EK N
Sbjct: 1486 KILEAQERLNQPSNINMEEIKKKWESEH---EQEVSQKIREAEEALKKRIRLPTEEKINK 1542
Query: 81 FFVEKEEEYVIKWKELQDKVAWAKSSDIELMTVGREIVDFHGEMVLLENYSALNYTGLVK 140
K+EE KE ++KV I+ M EI ++VL + A +
Sbjct: 1543 IIERKKEELE---KEFEEKV----EERIKSMEQSGEI-----DVVLRKQLEAKVQEKQKE 1590
Query: 141 IIKKYDKRTGALLR-LPFIQDVLNQPFFKI--DVLNKLVKECEVMLSIIFPKSGPLGQS- 196
+ +Y+K+ L+ +P + + K+ ++ ++L +E L I KS G+
Sbjct: 1591 LENEYNKKLQEELKDVPHSSHISDDERDKLRAEIESRLREEFNNELQAIKKKSFDEGKQQ 1650
Query: 197 --LSTSEVFEEVARETTTANETKETLDHVPKEFSEIQN 232
+ T+ + ++A+ + +ETK++ + PK + +QN
Sbjct: 1651 AMMKTTLLERKLAKMESQLSETKQSAESPPKSVNNVQN 1688
>LYS4_YEAST (P49367) Homoaconitase, mitochondrial precursor (EC
4.2.1.36) (Homoaconitate hydratase)
Length = 693
Score = 32.7 bits (73), Expect = 0.98
Identities = 19/58 (32%), Positives = 29/58 (49%)
Query: 171 VLNKLVKECEVMLSIIFPKSGPLGQSLSTSEVFEEVARETTTANETKETLDHVPKEFS 228
VL K+ EV+ + P SG + + V EEV +T ++ E L+ P+EFS
Sbjct: 456 VLGKISSPAEVLSTSEIPFSGVKTEIIENPVVEEEVNAQTEAPKQSVEILEGFPREFS 513
>KINH_HUMAN (P33176) Kinesin heavy chain (Ubiquitous kinesin heavy
chain) (UKHC)
Length = 963
Score = 32.3 bits (72), Expect = 1.3
Identities = 27/102 (26%), Positives = 45/102 (43%), Gaps = 4/102 (3%)
Query: 9 NQIEQTLPDWRDKFLSYKDLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFL 68
NQ Q + + + L ++L + ++ D+ ++ RL + A E KEV L
Sbjct: 440 NQQSQLVEKLKTQMLDQEELLASTR----RDQDNMQAELNRLQAENDASKEEVKEVLQAL 495
Query: 69 RLLEVEIEKFNGFFVEKEEEYVIKWKELQDKVAWAKSSDIEL 110
L V ++ + +K +EY + EL K A S D EL
Sbjct: 496 EELAVNYDQKSQEVEDKTKEYELLSDELNQKSATLASIDAEL 537
>Y832_METJA (Q58242) Hypothetical protein MJ0832 [Contains: Mja rnr-1
intein; Mja rnr-2 intein]
Length = 1750
Score = 32.0 bits (71), Expect = 1.7
Identities = 32/128 (25%), Positives = 58/128 (45%), Gaps = 10/128 (7%)
Query: 27 DLKKQLKLIVPKEIDSSCSKRRRLDDDGGAEGEVTKEVKDFLRLLEVEIEKFNGFFVEKE 86
DLK +I PK I S + L + + + ++KD ++ +E E E
Sbjct: 1173 DLKVGDFIITPKIIPSISKDKIYLSEIVKNKDKYYVKIKDHIKFIEEHEE----ILKESY 1228
Query: 87 EEYVIKWKELQDKVAWAKSSDIELMTVGREIVDFHGEMVLLENYSALNY-TGLVKIIKKY 145
+EY KWK+L+ + + ++L+ ++VD E + +Y NY +K+ +K+
Sbjct: 1229 KEYKTKWKDLKPVLKKKNAFRLDLI---EDLVD--KEKIEKISYGHANYINNKIKLDEKF 1283
Query: 146 DKRTGALL 153
GA L
Sbjct: 1284 GYLIGAFL 1291
>HMMR_RAT (P97779) Hyaluronan mediated motility receptor
(Intracellular hyaluronic acid binding protein)
(Receptor for hyaluronan-mediated motility)
Length = 498
Score = 32.0 bits (71), Expect = 1.7
Identities = 34/141 (24%), Positives = 69/141 (48%), Gaps = 16/141 (11%)
Query: 5 KILNNQIEQTLPDWRDKFLSYKDL-KKQLKLIVPKEIDSSCSKR-------RRLDDDGGA 56
K L+ ++Q L ++++ S +++ K+QLKL + E+D+ K ++L+++ +
Sbjct: 135 KELSAHLQQQLCSFQEEMTSERNVFKEQLKLALD-ELDAVQQKEEQSEKLVKQLEEETKS 193
Query: 57 EGEVTKEVKDFLRLLEVEIEKFNGFFVE----KEEEYVIKWKELQDKVAWAKSSDIELMT 112
E + + D LR E+E+EK + +E+Y + L+D A +S +
Sbjct: 194 TAEQLRRLDDLLREKEIELEKRTAAHAQATVIAQEKYSDTAQTLRDVTAQLESYKSSTL- 252
Query: 113 VGREIVDFHGEMVLLENYSAL 133
+EI D E + L+ A+
Sbjct: 253 --KEIEDLKLENLTLQEKVAM 271
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.136 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,994,862
Number of Sequences: 164201
Number of extensions: 1280824
Number of successful extensions: 4462
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 4417
Number of HSP's gapped (non-prelim): 71
length of query: 274
length of database: 59,974,054
effective HSP length: 109
effective length of query: 165
effective length of database: 42,076,145
effective search space: 6942563925
effective search space used: 6942563925
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC147471.4


