

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147471.11 - phase: 0 /pseudo
(1453 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
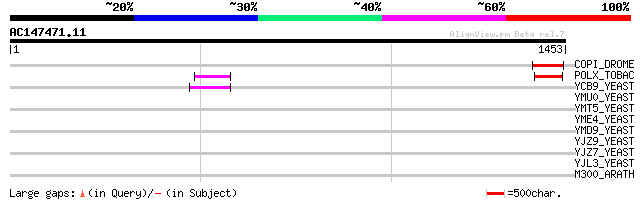
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 70 4e-11
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 61 3e-08
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 50 5e-05
YMU0_YEAST (Q04670) Transposon Ty1 protein B 45 0.001
YMT5_YEAST (Q04214) Transposon Ty1 protein B 45 0.001
YME4_YEAST (Q04711) Transposon Ty1 protein B 45 0.001
YMD9_YEAST (Q03434) Transposon Ty1 protein B 45 0.001
YJZ9_YEAST (P47100) Transposon Ty1 protein B 45 0.001
YJZ7_YEAST (P47098) Transposon Ty1 protein B 45 0.001
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 39 0.10
M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300... 39 0.10
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 70.1 bits (170), Expect = 4e-11
Identities = 29/79 (36%), Positives = 52/79 (65%)
Query: 1370 DEPMRLYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLL 1429
+ P+++Y DN+ I+IA+NP H R KH+++ HF +E++ + +IC Y+ +++ LAD+
Sbjct: 1323 ENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLADIF 1382
Query: 1430 TKRLNNNNFEKFVSKLGMI 1448
TK L F + KLG++
Sbjct: 1383 TKPLPAARFVELRDKLGLL 1401
Score = 36.6 bits (83), Expect = 0.52
Identities = 35/150 (23%), Positives = 59/150 (39%), Gaps = 18/150 (12%)
Query: 436 TKSRGGRLKLLENGMVFTIWITKTQAQSYLATTFFLESIKTNREKVLLYHYRVGDPSFRV 495
T S+ G + + +GM+ + + QA S A ++ L+H R G S
Sbjct: 379 TISKNGLMVVKNSGMLNNVPVINFQAYSINAK---------HKNNFRLWHERFGHISDGK 429
Query: 496 I-----KQLFP--LFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTF--PFYLVHTNVW 546
+ K +F L++ CE C K R+ F + P ++VH++V
Sbjct: 430 LLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQARLPFKQLKDKTHIKRPLFVVHSDVC 489
Query: 547 GPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
GP + +FV F+D T Y +
Sbjct: 490 GPITPVTLDDKNYFVIFVDQFTHYCVTYLI 519
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 60.8 bits (146), Expect = 3e-08
Identities = 28/73 (38%), Positives = 47/73 (64%)
Query: 1375 LYCDNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLN 1434
+YCD++ AI+++ N + H RTKH++V H+I+E +D + +S+ +N AD+LTK +
Sbjct: 1253 VYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVVP 1312
Query: 1435 NNNFEKFVSKLGM 1447
N FE +GM
Sbjct: 1313 RNKFELCKELVGM 1325
Score = 53.9 bits (128), Expect = 3e-06
Identities = 32/98 (32%), Positives = 52/98 (52%), Gaps = 6/98 (6%)
Query: 483 LYHYRVGDPSFR----VIKQLFPLFFKTLDVESFQCELCELAKHKRVTFSVSNKMSTFPF 538
L+H R+G S + + K+ + K V+ C+ C K RV+F S++
Sbjct: 424 LWHKRMGHMSEKGLQILAKKSLISYAKGTTVKP--CDYCLFGKQHRVSFQTSSERKLNIL 481
Query: 539 YLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFL 576
LV+++V GP + ++ G ++FVTFIDD + WVY L
Sbjct: 482 DLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYIL 519
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 50.1 bits (118), Expect = 5e-05
Identities = 35/120 (29%), Positives = 58/120 (48%), Gaps = 13/120 (10%)
Query: 472 ESIKTNREKVLLYHYRVGDPSFRVIKQLFP----LFFKTLDVE-----SFQCELCELAK- 521
+S N+ L H +G +FR I++ + K D+E ++QC C + K
Sbjct: 582 KSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCPDCLIGKS 641
Query: 522 --HKRVTFSVSNKMSTF-PFYLVHTNVWGPSNVPNISGARWFVTFIDDCTWVTWVYFLHD 578
H+ V S ++ PF +HT+++GP + S +F++F D+ T WVY LHD
Sbjct: 642 TKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHD 701
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 3/74 (4%)
Query: 507 LDVESFQCELCELAKHKRVTFSVSNKMSTF-PFYLVHTNVWGP-SNVPNISGARWFVTFI 564
+D + C + + KH+ + S +++ PF +HT+++GP N+P S +F++F
Sbjct: 206 IDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPK-SAPSYFISFT 264
Query: 565 DDCTWVTWVYFLHD 578
D+ T WVY LHD
Sbjct: 265 DETTKFRWVYPLHD 278
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 3/74 (4%)
Query: 507 LDVESFQCELCELAKHKRVTFSVSNKMSTF-PFYLVHTNVWGP-SNVPNISGARWFVTFI 564
+D + C + + KH+ + S +++ PF +HT+++GP N+P S +F++F
Sbjct: 206 IDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPK-SAPSYFISFT 264
Query: 565 DDCTWVTWVYFLHD 578
D+ T WVY LHD
Sbjct: 265 DETTKFRWVYPLHD 278
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 3/74 (4%)
Query: 507 LDVESFQCELCELAKHKRVTFSVSNKMSTF-PFYLVHTNVWGP-SNVPNISGARWFVTFI 564
+D + C + + KH+ + S +++ PF +HT+++GP N+P S +F++F
Sbjct: 206 IDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPK-SAPSYFISFT 264
Query: 565 DDCTWVTWVYFLHD 578
D+ T WVY LHD
Sbjct: 265 DETTKFRWVYPLHD 278
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 3/74 (4%)
Query: 507 LDVESFQCELCELAKHKRVTFSVSNKMSTF-PFYLVHTNVWGP-SNVPNISGARWFVTFI 564
+D + C + + KH+ + S +++ PF +HT+++GP N+P S +F++F
Sbjct: 206 IDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPK-SAPSYFISFT 264
Query: 565 DDCTWVTWVYFLHD 578
D+ T WVY LHD
Sbjct: 265 DETTKFRWVYPLHD 278
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 3/74 (4%)
Query: 507 LDVESFQCELCELAKHKRVTFSVSNKMSTF-PFYLVHTNVWGP-SNVPNISGARWFVTFI 564
+D + C + + KH+ + S +++ PF +HT+++GP N+P S +F++F
Sbjct: 633 IDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPK-SAPSYFISFT 691
Query: 565 DDCTWVTWVYFLHD 578
D+ T WVY LHD
Sbjct: 692 DETTKFRWVYPLHD 705
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 45.4 bits (106), Expect = 0.001
Identities = 24/74 (32%), Positives = 42/74 (56%), Gaps = 3/74 (4%)
Query: 507 LDVESFQCELCELAKHKRVTFSVSNKMSTF-PFYLVHTNVWGP-SNVPNISGARWFVTFI 564
+D + C + + KH+ + S +++ PF +HT+++GP N+P S +F++F
Sbjct: 633 IDYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPK-SAPSYFISFT 691
Query: 565 DDCTWVTWVYFLHD 578
D+ T WVY LHD
Sbjct: 692 DETTKFRWVYPLHD 705
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 38.9 bits (89), Expect = 0.10
Identities = 21/68 (30%), Positives = 37/68 (53%)
Query: 1378 DNKYAINIAHNPVQHDRTKHLEVNKHFIKEKLDSGLICTPYVSSQDNLADLLTKRLNNNN 1437
D+K AI + Q + K + IKEK+ I ++ + N+ADLLTK ++ ++
Sbjct: 1721 DSKPAIQGLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLKITGKGNIADLLTKPVSASD 1780
Query: 1438 FEKFVSKL 1445
F++F+ L
Sbjct: 1781 FKRFIQVL 1788
>M300_ARATH (P93293) Hypothetical mitochondrial protein AtMg00300
(ORF145a) (ORF1451)
Length = 145
Score = 38.9 bits (89), Expect = 0.10
Identities = 25/80 (31%), Positives = 40/80 (49%), Gaps = 2/80 (2%)
Query: 475 KTNREKVLLYHYRVGDPSFRVIKQLFPL-FFKTLDVESFQ-CELCELAKHKRVTFSVSNK 532
+T +++ L+H R+ S R ++ L F + V S + CE C K RV FS
Sbjct: 63 ETAKDETRLWHSRLAHMSQRGMELLVKKGFLDSSKVSSLKFCEDCIYGKTHRVNFSTGQH 122
Query: 533 MSTFPFYLVHTNVWGPSNVP 552
+ P VH+++WG +VP
Sbjct: 123 TTKNPLDYVHSDLWGAPSVP 142
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.364 0.163 0.626
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 137,913,688
Number of Sequences: 164201
Number of extensions: 4855306
Number of successful extensions: 24382
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 24355
Number of HSP's gapped (non-prelim): 27
length of query: 1453
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1330
effective length of database: 39,777,331
effective search space: 52903850230
effective search space used: 52903850230
T: 11
A: 40
X1: 14 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 36 (21.5 bits)
S2: 72 (32.3 bits)
Medicago: description of AC147471.11


