

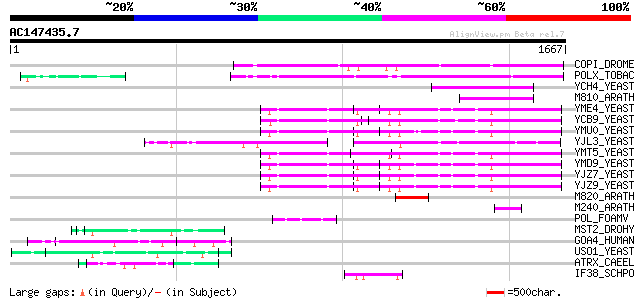
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.7 + phase: 0
(1667 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 516 e-145
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 506 e-142
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 168 1e-40
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 157 2e-37
YME4_YEAST (Q04711) Transposon Ty1 protein B 127 2e-28
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 127 2e-28
YMU0_YEAST (Q04670) Transposon Ty1 protein B 125 1e-27
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 124 2e-27
YMT5_YEAST (Q04214) Transposon Ty1 protein B 123 4e-27
YMD9_YEAST (Q03434) Transposon Ty1 protein B 123 4e-27
YJZ7_YEAST (P47098) Transposon Ty1 protein B 121 1e-26
YJZ9_YEAST (P47100) Transposon Ty1 protein B 121 2e-26
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 110 3e-23
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 63 6e-09
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 60 5e-08
MST2_DROHY (Q08696) Axoneme-associated protein mst101(2) 60 5e-08
GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member... 60 5e-08
USO1_YEAST (P25386) Intracellular protein transport protein USO1 59 9e-08
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 57 4e-07
IF38_SCHPO (O14164) Probable eukaryotic translation initiation f... 55 1e-06
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 516 bits (1328), Expect = e-145
Identities = 338/1079 (31%), Positives = 553/1079 (50%), Gaps = 108/1079 (10%)
Query: 671 ISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGKRVENVYKIN 730
I++ +V NL+S+ + + G + F K+ T ++K+ + + NV IN
Sbjct: 343 ITLEDVLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVT-ISKNGLMVVKNSGMLNNVPVIN 401
Query: 731 FSDLADQKVVCLLSMNDKKW----VWHKRLGHANWRLISKISKLQLVKG---LPNIDYHS 783
F S+N K +WH+R GH + + +I + + L N++
Sbjct: 402 FQ---------AYSINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSC 452
Query: 784 DALCGACQKGKIVKSSFKS-KDIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYS 842
+ +C C GK + FK KD RPL ++H D+ GP+ +L Y ++ VD ++
Sbjct: 453 E-ICEPCLNGKQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFT 511
Query: 843 RWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILH 902
+ IK K +F F + ++ LK++ + D+G E+ + FC K GI +
Sbjct: 512 HYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISY 571
Query: 903 EFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLE 962
+ P TPQ NGV ER RT+ E ARTM+ L K FW EAV T+ Y+ NRI R +++
Sbjct: 572 HLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVD 631
Query: 963 --KTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERS------ 1014
KT YE++ ++P + + FG T Y+ + K+ KFD K+ + IF+GY
Sbjct: 632 SSKTPYEMWHNKKPYLKHLRVFGATVYV-HIKNKQGKFDDKSFKSIFVGYEPNGFKLWDA 690
Query: 1015 ------KAYRVYNSETQCVEESMHVKF------DDREPESKT----------------SE 1046
A V ET V S VKF D +E E+K S+
Sbjct: 691 VNEKFIVARDVVVDETNMVN-SRAVKFETVFLKDSKESENKNFPNDSRKIIQTEFPNESK 749
Query: 1047 QSESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEITPEAESNSEAEPSPKV-QNESAS 1105
+ ++ DS+++ + P+DS K + E E++ + +++ S K NES
Sbjct: 750 ECDNIQFLKDSKESENKNFPNDSRKIIQTEFPNESKECDNIQFLKDSKESNKYFLNESKK 809
Query: 1106 ---EDFQDNTQQVIQPKFKHKSSHHEEL---------------IIGSKDSPRRTRSHFRQ 1147
+D + ++ P +S E L II + +T+
Sbjct: 810 RKRDDHLNESKGSGNPNESRESETAEHLKEIGIDNPTKNDGIEIINRRSERLKTKPQISY 869
Query: 1148 EESLIGLLSII---------EPKTVEEALSDD---GWILAMQEELNQFQRNDVWDLVPKP 1195
E L ++ P + +E D W A+ ELN + N+ W + +P
Sbjct: 870 NEEDNSLNKVVLNAHTIFNDVPNSFDEIQYRDDKSSWEEAINTELNAHKINNTWTITKRP 929
Query: 1196 FQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLS 1255
KNI+ ++WVF K NE G R KARLVA+G++Q+ IDY ETFAPVAR+ + R +LS
Sbjct: 930 ENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETFAPVARISSFRFILS 989
Query: 1256 YAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRA 1315
I + + ++QMDVK+AFLNG ++EE+Y++ P G + ++V KL K++YGLKQA R
Sbjct: 990 LVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGIS--CNSDNVCKLNKAIYGLKQAARC 1047
Query: 1316 WYDRLSNFLIKNDFERGQVDTTLF---RRTLKKDILIVQIYVDDIIFGSTNASLCKEFSK 1372
W++ L + +F VD ++ + + ++I ++ +YVDD++ + + + F +
Sbjct: 1048 WFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYVL-LYVDDVVIATGDMTRMNNFKR 1106
Query: 1373 LMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTC 1432
+ ++F M+ + E+K F+GI+I ++ +Y+ Q+ Y K++L KF +E+C ++TP+
Sbjct: 1107 YLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMENCNAVSTPLPSKI 1166
Query: 1433 TL----SKEDTGTVVDQKLYRGMIGSLLY-LTASRPDILFSVCLCARFQSDPRESHLTAV 1487
S ED T R +IG L+Y + +RPD+ +V + +R+ S +
Sbjct: 1167 NYELLNSDEDCNTPC-----RSLIGCLMYIMLCTRPDLTTAVNILSRYSSKNNSELWQNL 1221
Query: 1488 KRIFRYLKGTTNLGLLYRKSLDY--KLIGFCDADYAGDRIERKSTSGNC-QFLGENLISW 1544
KR+ RYLKGT ++ L+++K+L + K+IG+ D+D+AG I+RKST+G + NLI W
Sbjct: 1222 KRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKMFDFNLICW 1281
Query: 1545 ASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQIN-ANSIPIYCDNTAAICLSKN 1603
+K+Q ++A S+ EAEY++ + LW+K L I N I IY DN I ++ N
Sbjct: 1282 NTKRQNSVAASSTEAEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCISIANN 1341
Query: 1604 PILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKKNLNM 1662
P H RAKHI+IK+HF R+ VQ ++ +++I TE+Q ADIFTKPL RF ++ L +
Sbjct: 1342 PSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTENQLADIFTKPLPAARFVELRDKLGL 1400
Score = 34.7 bits (78), Expect = 2.3
Identities = 47/211 (22%), Positives = 80/211 (37%), Gaps = 43/211 (20%)
Query: 119 TAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSGLQILK 178
TA+ + +L A +E + A L L + ++ + S+ + F L+S L
Sbjct: 76 TARQILENLDAVYE---RKSLASQLALRKRLLSLKLSSEMSLLSHFHIFDELISELLAAG 132
Query: 179 KSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSL-NEH-ETS 236
D +S +L +LPS + +TAIE + N L++ + + L E+ + N+H +TS
Sbjct: 133 AKIEEMDKISHLLITLPSCYDGIITAIETLSEEN-LTLAFVKNRLLDQEIKIKNDHNDTS 191
Query: 237 KKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKF 296
KK + + + T K +N + K KK
Sbjct: 192 KKVMNAIVHNNNNTYK------------------------------NNLFKNRVTKPKKI 221
Query: 297 LSKRGSYKNSKKEDQKGCFNCKKPGHFIVDC 327
YK C +C + GH DC
Sbjct: 222 FKGNSKYKVK-------CHHCGREGHIKKDC 245
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 506 bits (1304), Expect = e-142
Identities = 332/1025 (32%), Positives = 541/1025 (52%), Gaps = 63/1025 (6%)
Query: 663 KSTIGNSSISINNVWLVDGLKHNLLSISQFCDNGYDVTFSKTNCTLVNKDDKSITFKGKR 722
K+ +G ++ + +V V L+ NL+S +GY+ F+ L + KG
Sbjct: 341 KTNVG-CTLVLKDVRHVPDLRMNLISGIALDRDGYESYFANQKWRLTK--GSLVIAKGVA 397
Query: 723 VENVYKINFSDLADQKVVCLLSMNDKKW-----VWHKRLGHANWRLISKISKLQLVKGLP 777
+Y+ N +C +N + +WHKR+GH + + + ++K L+
Sbjct: 398 RGTLYRTNAE-------ICQGELNAAQDEISVDLWHKRMGHMSEKGLQILAKKSLISYAK 450
Query: 778 NIDYHSDALCGACQKGKIVKSSFKSKDIVSTSRPLELLHI---DLFGPVNTASLYGSKYG 834
C C GK + SF++ S+ R L +L + D+ GP+ S+ G+KY
Sbjct: 451 GTTVKP---CDYCLFGKQHRVSFQT----SSERKLNILDLVYSDVCGPMEIESMGGNKYF 503
Query: 835 LVIVDDYSRWTWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGGEFENEPFELF 894
+ +DD SR WV +K+KD +VF F ++ E K+ ++RSD+GGE+ + FE +
Sbjct: 504 VTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEY 563
Query: 895 CEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNR 954
C HGI HE + P TPQ NGV ER NRT+ E R+M+ L K FW EAV T+CY+ NR
Sbjct: 564 CSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINR 623
Query: 955 IYIRPMLEKTAYELFKGRRPNISYFHQFGCTCYILNTKDYLKKFDAKAQRGIFLGYSERS 1014
P+ + ++ + + S+ FGC + K+ K D K+ IF+GY +
Sbjct: 624 SPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEE 683
Query: 1015 KAYRVYNSETQCVEESMHVKFDDREPESKT----SEQSESNAGTTDSEDASESDQPSDSE 1070
YR+++ + V S V F RE E +T SE+ ++ S S+ P+ +E
Sbjct: 684 FGYRLWDPVKKKVIRSRDVVF--RESEVRTAADMSEKVKNGIIPNFVTIPSTSNNPTSAE 741
Query: 1071 KYTEVESSPEAEITPEAESNSEA--EPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHE 1128
T+ E S + E E E E +V++ + E+ ++ +P+ + +
Sbjct: 742 STTD-EVSEQGEQPGEVIEQGEQLDEGVEEVEHPTQGEEQHQPLRRSERPRVESRRYPST 800
Query: 1129 ELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDV 1188
E ++ S D R+ ESL +LS E + + AMQEE+ Q+N
Sbjct: 801 EYVLISDD---------REPESLKEVLSHPEKNQL---------MKAMQEEMESLQKNGT 842
Query: 1189 WDLVPKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLE 1248
+ LV P K + KWVF+ K + ++ R KARLV +G+ Q++GID+ E F+PV ++
Sbjct: 843 YKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMT 902
Query: 1249 AIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYG 1308
+IR +LS A + + + Q+DVK+AFL+G +EEE+Y++QP GFE + V KL KSLYG
Sbjct: 903 SIRTILSLAASLDLEVEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYG 962
Query: 1309 LKQAPRAWYDRLSNFLIKNDFERGQVDTTL-FRRTLKKDILIVQIYVDDIIFGSTNASLC 1367
LKQAPR WY + +F+ + + D + F+R + + +I+ +YVDD++ + L
Sbjct: 963 LKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLI 1022
Query: 1368 KEFSKLMQDEFEMSMMGELKFFLGIQI--NQSKEGVYVHQTKYTKELLKKFKLEDCKVMN 1425
+ + F+M +G + LG++I ++ +++ Q KY + +L++F +++ K ++
Sbjct: 1023 AKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVS 1082
Query: 1426 TPMHPTCTLSKEDTGTVVDQK------LYRGMIGSLLY-LTASRPDILFSVCLCARFQSD 1478
TP+ LSK+ T V++K Y +GSL+Y + +RPDI +V + +RF +
Sbjct: 1083 TPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHAVGVVSRFLEN 1142
Query: 1479 PRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLG 1538
P + H AVK I RYL+GTT L + S D L G+ DAD AGD RKS++G
Sbjct: 1143 PGKEHWEAVKWILRYLRGTTGDCLCFGGS-DPILKGYTDADMAGDIDNRKSSTGYLFTFS 1201
Query: 1539 ENLISWASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTAAI 1598
ISW SK Q +A+ST EAEYI+A +++W+K L++ ++ +YCD+ +AI
Sbjct: 1202 GGAISWQSKLQKCVALSTTEAEYIAATETGKEMIWLKRFLQELGLHQKEYVVYCDSQSAI 1261
Query: 1599 CLSKNPILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFIKK 1658
LSKN + H+R KHI++++H+IR+ V L + I T AD+ TK + +F+ K+
Sbjct: 1262 DLSKNSMYHARTKHIDVRYHWIREMVDDESLKVLKISTNENPADMLTKVVPRNKFELCKE 1321
Query: 1659 NLNMH 1663
+ MH
Sbjct: 1322 LVGMH 1326
Score = 56.6 bits (135), Expect = 6e-07
Identities = 72/324 (22%), Positives = 122/324 (37%), Gaps = 77/324 (23%)
Query: 33 KFNGDPEEFSWWKTNMYSF--------IMGLDEELWDILEDGVDDLDLDEEGAAIDRRIH 84
KFNGD FS W+ M ++ +D + D ++ D DLDE A+ R
Sbjct: 10 KFNGD-NGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMK-AEDWADLDERAASAIR--- 64
Query: 85 TPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAMFASLCANFEGSKKVKEAKALM 144
L+ + II D+ TA+ ++ L + + + L
Sbjct: 65 ------LHLSDDVVNNII-------------DEDTARGIWTRLESLYMSKTLTNK---LY 102
Query: 145 LVHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTA 204
L Q M + + + F L++ L L D +L SLPS + T
Sbjct: 103 LKKQLYALHMSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATT 162
Query: 205 IEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKT-SKSSKAYKASESE 263
I K T+ ++D+ S+L ++E + E + +++ +G++ +SS Y S +
Sbjct: 163 ILHGK--TTIELKDVTSALLLNEKMRKKPEN--QGQALITEGRGRSYQRSSNNYGRSGA- 217
Query: 264 EESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHF 323
RG KN K + C+NC +PGHF
Sbjct: 218 ------------------------------------RGKSKNRSKSRVRNCYNCNQPGHF 241
Query: 324 IVDCPDLQKEKFKGKSMKSSFNSS 347
DCP+ +K K + K+ N++
Sbjct: 242 KRDCPNPRKGKGETSGQKNDDNTA 265
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 168 bits (425), Expect = 1e-40
Identities = 96/308 (31%), Positives = 167/308 (54%), Gaps = 3/308 (0%)
Query: 1267 MDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLIK 1326
MDV +AFLN ++E +YVKQPPGF + ++P++V++L +YGLKQAP W + ++N L K
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1327 NDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGEL 1386
F R + + L+ R+ + + +YVDD++ + + + + + + M +G++
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 1387 KFFLGIQINQSKEG-VYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQ 1445
FLG+ I+QS G + + Y + + ++ K+ TP+ + L + + + D
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 1446 KLYRGMIGSLLY-LTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLY 1504
Y+ ++G LL+ RPDI + V L +RF +PR HL + +R+ RYL T ++ L Y
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 1505 RKSLDYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKK-QATIAMSTAEAEYIS 1563
R L +CDA + ST G L ++W+SKK + I + + EAEYI+
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 1564 AANCCTQL 1571
A+ ++
Sbjct: 301 ASETVMEI 308
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 157 bits (397), Expect = 2e-37
Identities = 81/222 (36%), Positives = 128/222 (57%), Gaps = 1/222 (0%)
Query: 1352 IYVDDIIFGSTNASLCKEFSKLMQDEFEMSMMGELKFFLGIQINQSKEGVYVHQTKYTKE 1411
+YVDDI+ ++ +L + F M +G + +FLGIQI G+++ QTKY ++
Sbjct: 5 LYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYAEQ 64
Query: 1412 LLKKFKLEDCKVMNTPMHPTCTLSKEDTGTVVDQKLYRGMIGSLLYLTASRPDILFSVCL 1471
+L + DCK M+TP+ P S T D +R ++G+L YLT +RPDI ++V +
Sbjct: 65 ILNNAGMLDCKPMSTPL-PLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAVNI 123
Query: 1472 CARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGFCDADYAGDRIERKSTS 1531
+ +P + +KR+ RY+KGT GL K+ + FCD+D+AG R+ST+
Sbjct: 124 VCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRSTT 183
Query: 1532 GNCQFLGENLISWASKKQATIAMSTAEAEYISAANCCTQLLW 1573
G C FLG N+ISW++K+Q T++ S+ E EY + A +L W
Sbjct: 184 GFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 127 bits (320), Expect = 2e-28
Identities = 159/688 (23%), Positives = 297/688 (43%), Gaps = 82/688 (11%)
Query: 1034 KFDDREPESKTSEQSESNAGTTDSEDASESDQP-----SDSEKYTE---VESSPEAEITP 1085
+ D+E E + +S S + ++S + P + SE+ TE + P ++ P
Sbjct: 652 QISDQETEKRIIHRSPSIDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPP 711
Query: 1086 EAESN-----SEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDS--- 1137
E+ + E P Q S+ D+ K +E I S+D+
Sbjct: 712 ESPTEFPDPFKELPPINSHQTNSSLGGIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNT 771
Query: 1138 -------PRRTRSHFRQEESLIGLLSIIEPKTV---EEALS-------DDGWILAMQEEL 1180
P R++ ++ + SI +T +EA++ + +I A +E+
Sbjct: 772 KNMRSLEPPRSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEV 831
Query: 1181 NQFQRNDVWDLVPKPFQKNIIGTKWVFRNKL--NEQGEVTRNKARLVAQGYSQQEGIDYT 1238
NQ + + WD K + + I K V + N + + T +KAR VA+G Q +
Sbjct: 832 NQLLKMNTWD-TDKYYDRKEIDPKRVINSMFIFNRKRDGT-HKARFVARGDIQHPDTYDS 889
Query: 1239 ETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNH 1298
+ A+ LS A+++ + Q+D+ SA+L I+EE+Y++ PP L +
Sbjct: 890 GMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP---HLGMNDK 946
Query: 1299 VYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDII 1358
+ +LKKSLYGLKQ+ WY+ + ++LIK + G + + K + + ++VDD+I
Sbjct: 947 LIRLKKSLYGLKQSGANWYETIKSYLIK---QCGMEEVRGWSCVFKNSQVTICLFVDDMI 1003
Query: 1359 FGSTNASLCKEFSKLMQDEFEMSMM------GELKF-FLGIQINQSKEGVYVHQTKYTKE 1411
S + + K+ ++ +++ ++ E+++ LG++I + KY K
Sbjct: 1004 LFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQR-------GKYMKL 1056
Query: 1412 LLKKFKLEDCKVMNTPMHP--TCTLSKEDTGTVVDQ------------KLY--RGMIGSL 1455
++ E +N P++P + G +DQ K++ + +IG
Sbjct: 1057 GMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLA 1116
Query: 1456 LYLTAS-RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSL----DY 1510
Y+ R D+L+ + A+ P L + +++ T + L++ K+ D
Sbjct: 1117 SYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDN 1176
Query: 1511 KLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAANCCTQ 1570
KL+ DA Y G++ KS GN L +I S K + ST EAE + +
Sbjct: 1177 KLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPL 1235
Query: 1571 LLWMKHQLEDYQINANSIPIYCD--NTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKGI 1628
L + H +++ + + D +T +I +S N R + K +RD V
Sbjct: 1236 LNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNE-EKFRNRFFGTKAMRLRDEVSGNH 1294
Query: 1629 LDIQFIDTEHQWADIFTKPLSVERFDFI 1656
L + +I+T+ AD+ TKPL ++ F +
Sbjct: 1295 LHVCYIETKKNIADVMTKPLPIKTFKLL 1322
Score = 95.5 bits (236), Expect = 1e-18
Identities = 90/388 (23%), Positives = 166/388 (42%), Gaps = 34/388 (8%)
Query: 753 HKRLGHANWRLISKISKLQLVKGLPN--------IDYHS-DALCGACQKGKIVKSSFKSK 803
H+ L HAN + I K + IDY D L G K + +K S
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKY 231
Query: 804 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFS 861
++ P + LH D+FGPV+ Y + D+ +++ WV + + +D +VF+
Sbjct: 232 Q--NSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFT 289
Query: 862 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 921
+ I+++ + +L ++ D G E+ N F EK+GI +++ + +GV ER NR
Sbjct: 290 TILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNR 349
Query: 922 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 981
TL + RT + + L H W A+ S ++N + P +K+A + +IS
Sbjct: 350 TLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLP 408
Query: 982 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETQCVEESMHVKFDDREP 1040
FG I+N + K + G L S S Y +Y S + V+ + +V +E
Sbjct: 409 FG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKES 467
Query: 1041 E---------------SKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPE--AEI 1083
++ + +S + + + + + + SD + +++E PE +
Sbjct: 468 RLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNV 527
Query: 1084 TPEAESNSEAEPSPKVQNESASEDFQDN 1111
+A S +++ P P E + + N
Sbjct: 528 LSKAVSPTDSTP-PSTHTEDSKRVSKTN 554
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 127 bits (320), Expect = 2e-28
Identities = 152/661 (22%), Positives = 292/661 (43%), Gaps = 78/661 (11%)
Query: 1057 SEDASESDQPSDSEKYTEVESSPEAEITPEAESNSEAEPS------PKVQNESASEDF-- 1108
S+ A+ + P S K +S + P+ S + S P + + +
Sbjct: 1121 SDHATPNIMPDKSSKNVTADSILDDLPLPDLTHQSPTDTSDVSKDIPHIHSRQTNSSLGG 1180
Query: 1109 QDNTQQVIQPKFKHKSSHHEELIIG-SKDS----------PRRTRSHFRQEESLIGLLSI 1157
D++ + K K +S E I S+D+ P R++ ++ G+ SI
Sbjct: 1181 MDDSNVLTTTKSKKRSLEDNETEIEVSRDTWNNKNMRSLEPPRSKKRINLIAAIKGVKSI 1240
Query: 1158 IEPKTV---EEALS-------DDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTKWVF 1207
+T +EA++ D ++ A +E++Q + + WD K + +N I K V
Sbjct: 1241 KPVRTTLRYDEAITYNKDNKEKDRYVEAYHKEISQLLKMNTWD-TNKYYDRNDIDPKKVI 1299
Query: 1208 RNKL--NEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHGIILY 1265
+ N++ + T +KAR VA+G Q ++ + A+ LS A+++ +
Sbjct: 1300 NSMFIFNKKRDGT-HKARFVARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYIT 1358
Query: 1266 QMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLI 1325
Q+D+ SA+L I+EE+Y++ PP L + + +L+KSLYGLKQ+ WY+ + ++LI
Sbjct: 1359 QLDISSAYLYADIKEELYIRPPP---HLGLNDKLLRLRKSLYGLKQSGANWYETIKSYLI 1415
Query: 1326 K-NDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMM- 1383
D + + + +F K + + ++VDD+I S + + K+ ++ +++ ++
Sbjct: 1416 NCCDMQEVRGWSCVF----KNSQVTICLFVDDMILFSKDLNANKKIITTLKKQYDTKIIN 1471
Query: 1384 -----GELKF-FLGIQINQSKEGVYVHQTKYTKELLKKFKLEDCKVMNTPMHPTCTLSKE 1437
E+++ LG++I + +KY K ++K E +N P++P +
Sbjct: 1472 LGESDNEIQYDILGLEIKYQR-------SKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRA 1524
Query: 1438 --DTGTVVDQ------------KLY--RGMIGSLLYLTAS-RPDILFSVCLCARFQSDPR 1480
G +DQ K++ + +IG Y+ R D+L+ + A+ P
Sbjct: 1525 PGQPGHYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPS 1584
Query: 1481 ESHLTAVKRIFRYLKGTTNLGLLYRKSL----DYKLIGFCDADYAGDRIERKSTSGNCQF 1536
L + +++ T + L++ K+ D KL+ DA Y G++ KS GN
Sbjct: 1585 RQVLDMTYELIQFMWDTRDKQLIWHKNKPTKPDNKLVAISDASY-GNQPYYKSQIGNIFL 1643
Query: 1537 LGENLISWASKKQATIAMSTAEAEYISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTA 1596
L +I S K + ST EAE + + L + H +++ + D+ +
Sbjct: 1644 LNGKVIGGKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPIIKGLLTDSRS 1703
Query: 1597 AICLSKNPILHS-RAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDF 1655
I + K+ R + K +RD V L + +I+T+ AD+ TKPL ++ F
Sbjct: 1704 TISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKL 1763
Query: 1656 I 1656
+
Sbjct: 1764 L 1764
Score = 105 bits (261), Expect = 1e-21
Identities = 89/351 (25%), Positives = 154/351 (43%), Gaps = 31/351 (8%)
Query: 753 HKRLGHANWRLISKISKLQLVKGLPNIDYH---------SDALCGACQKGKIVKSSFKSK 803
H+ LGHAN+R I K K V L D D L G K + VK S
Sbjct: 595 HRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCPDCLIGKSTKHRHVKGSRLKY 654
Query: 804 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFS 861
+ P + LH D+FGPV+ Y + D+ +R+ WV + + ++ VF+
Sbjct: 655 Q--ESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFT 712
Query: 862 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 921
S I+++ ++L ++ D G E+ N+ F GI +++ + +GV ER NR
Sbjct: 713 SILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNR 772
Query: 922 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 981
TL RT++H + L H W AV S I+N + + P +K+A + +I+
Sbjct: 773 TLLNDCRTLLHCSGLPNHLWFSAVEFSTIIRNSL-VSPKNDKSARQHAGLAGLDITTILP 831
Query: 982 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETQCVEESMHVKFDDREP 1040
FG I+N + K + G L S S Y +Y S + V+ + +V D++
Sbjct: 832 FG-QPVIVNNHNPDSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQDKQS 890
Query: 1041 E---------------SKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVE 1076
+ ++ + ++S ++E + + + SD + +E+E
Sbjct: 891 KLDQFNYDTLTFDDDLNRLTAHNQSFIEQNETEQSYDQNTESDHDYQSEIE 941
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 125 bits (314), Expect = 1e-27
Identities = 158/696 (22%), Positives = 297/696 (41%), Gaps = 98/696 (14%)
Query: 1034 KFDDREPESKTSEQSESNAGTTDSEDASESDQP-----SDSEKYTE---VESSPEAEITP 1085
+ D+E E + +S S + ++S + P + SE+ TE + P ++ P
Sbjct: 652 QISDQETEKRIIHRSPSIDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPP 711
Query: 1086 EAESN-----SEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDS--- 1137
E+ + E P Q S+ D+ K +E I S+D+
Sbjct: 712 ESPTEFPDPFKELPPINSRQTNSSLGGIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNT 771
Query: 1138 -------PRRTRSHFRQEESLIGLLSIIEPKTV---EEALS-------DDGWILAMQEEL 1180
P R++ ++ + SI +T +EA++ + +I A +E+
Sbjct: 772 KNMRSLEPPRSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIQAYHKEV 831
Query: 1181 NQFQRNDVWDLV-----PKPFQKNIIGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGI 1235
NQ + WD + K +I + ++F K + +KAR VA+G I
Sbjct: 832 NQLLKMKTWDTDRYYDRKEIDPKRVINSMFIFNRKRDGT-----HKARFVARG-----DI 881
Query: 1236 DYTETFAPVARLE-----AIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGF 1290
+ +T+ P + A+ LS A+++ + Q+D+ SA+L I+EE+Y++ PP
Sbjct: 882 QHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP-- 939
Query: 1291 EDLKHPNHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIV 1350
L + + +LKKSLYGLKQ+ WY+ + ++LIK + G + + K + +
Sbjct: 940 -HLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIK---QCGMEEVRGWSCVFKNSQVTI 995
Query: 1351 QIYVDDIIFGSTNASLCKEFSKLMQDEFEMSMM------GELKF-FLGIQINQSKEGVYV 1403
++VDD+I S + + K+ ++ +++ ++ E+++ LG++I +
Sbjct: 996 CLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQR----- 1050
Query: 1404 HQTKYTKELLKKFKLEDCKVMNTPMHP--TCTLSKEDTGTVVDQK--------------L 1447
KY K ++ E +N P++P + G +DQ+
Sbjct: 1051 --GKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHE 1108
Query: 1448 YRGMIGSLLYLTAS-RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRK 1506
+ +IG Y+ R D+L+ + A+ P + L + +++ T + L++ K
Sbjct: 1109 MQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHK 1168
Query: 1507 SLDY----KLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYI 1562
S KL+ DA Y G++ KS GN L +I S K + ST EAE
Sbjct: 1169 SKPVKPTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIH 1227
Query: 1563 SAANCCTQLLWMKHQLEDYQINANSIPIYCD--NTAAICLSKNPILHSRAKHIEIKHHFI 1620
+ + L + H +++ + + D +T +I +S N R + K +
Sbjct: 1228 AISESVPLLNNLSHLVQELNKKPITKGLLTDSKSTISIIISNNE-EKFRNRFFGTKAMRL 1286
Query: 1621 RDYVQKGILDIQFIDTEHQWADIFTKPLSVERFDFI 1656
RD V L + +I+T+ AD+ TKPL ++ F +
Sbjct: 1287 RDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLL 1322
Score = 95.5 bits (236), Expect = 1e-18
Identities = 90/388 (23%), Positives = 166/388 (42%), Gaps = 34/388 (8%)
Query: 753 HKRLGHANWRLISKISKLQLVKGLPN--------IDYHS-DALCGACQKGKIVKSSFKSK 803
H+ L HAN + I K + IDY D L G K + +K S
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKY 231
Query: 804 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFS 861
++ P + LH D+FGPV+ Y + D+ +++ WV + + +D +VF+
Sbjct: 232 Q--NSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFT 289
Query: 862 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 921
+ I+++ + +L ++ D G E+ N F EK+GI +++ + +GV ER NR
Sbjct: 290 TILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNR 349
Query: 922 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 981
TL + RT + + L H W A+ S ++N + P +K+A + +IS
Sbjct: 350 TLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLP 408
Query: 982 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETQCVEESMHVKFDDREP 1040
FG I+N + K + G L S S Y +Y S + V+ + +V +E
Sbjct: 409 FG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKES 467
Query: 1041 E---------------SKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPE--AEI 1083
++ + +S + + + + + + SD + +++E PE +
Sbjct: 468 RLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNV 527
Query: 1084 TPEAESNSEAEPSPKVQNESASEDFQDN 1111
+A S +++ P P E + + N
Sbjct: 528 LSKAVSPTDSTP-PSTHTEDSKRVSKTN 554
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 124 bits (311), Expect = 2e-27
Identities = 152/657 (23%), Positives = 274/657 (41%), Gaps = 51/657 (7%)
Query: 1034 KFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTE--VESSPEAEITPEAESNS 1091
KF+ E + TD E + K T+ V + ++P E N
Sbjct: 1139 KFEKENHHPPPIEDIVDMSDQTDMESNCQDGNNLKELKVTDKNVPTDNGTNVSPRLEQNI 1198
Query: 1092 EAEPSP-KVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSHFRQEES 1150
EA SP + N+SA + + ++ + + + +H ++ +D R ++ +
Sbjct: 1199 EASGSPVQTVNKSAFLNKEFSSLNMKRKRKRHDKNNSLTSYELERDKKRSKKNRVKLIPD 1258
Query: 1151 LIGLLSIIEPKTV--EEALSDDG-------WILAMQEELNQFQRNDVWDLVPKPFQKNII 1201
+ +S + + + EA+S + + A +EL + V+D+ K + I
Sbjct: 1259 NMETVSAPKIRAIYYNEAISKNPDLKEKHEYKQAYHKELQNLKDMKVFDVDVKYSRSEIP 1318
Query: 1202 GTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYAINHG 1261
V N + + KAR+V +G +Q Y+ I++ L A N
Sbjct: 1319 DNLIVPTNTIFTKKRNGIYKARIVCRGDTQSPDT-YSVITTESLNHNHIKIFLMIANNRN 1377
Query: 1262 IILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPNH-VYKLKKSLYGLKQAPRAWYDRL 1320
+ + +D+ AFL +EEE+Y+ P H V KL K+LYGLKQ+P+ W D L
Sbjct: 1378 MFMKTLDINHAFLYAKLEEEIYIPHP-------HDRRCVVKLNKALYGLKQSPKEWNDHL 1430
Query: 1321 SNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDIIFGSTNASLCKEFSKLMQDEFEM 1380
+L + L++ K L++ +YVDD + ++N EF ++ FE+
Sbjct: 1431 RQYLNGIGLKDNSYTPGLYQTEDKN--LMIAVYVDDCVIAASNEQRLDEFINKLKSNFEL 1488
Query: 1381 SMMGEL------KFFLGIQINQSKEGVYVHQT--KYTKELLKKFKLEDCKVMNTPMHPTC 1432
+ G L LG+ + +K + T + + KK+ E K+ + +
Sbjct: 1489 KITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMS 1548
Query: 1433 TLS---KEDTGTVVDQKLYRG------MIGSLLYLT-ASRPDILFSVCLCARFQSDPRES 1482
T K+D + +++ +G ++G L Y+ R DI F+V AR + P E
Sbjct: 1549 TYKIDPKKDVLQMSEEEFRQGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHER 1608
Query: 1483 HLTAVKRIFRYLKGTTNLGLLYRK--SLDYKLIGFCDADYAGDRIERKSTSGNCQFLGEN 1540
+ +I +YL ++G+ Y + + D K+I DA G + +S G + G N
Sbjct: 1609 VFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIAITDAS-VGSEYDAQSRIGVILWYGMN 1667
Query: 1541 LISWASKKQATIAMSTAEAE----YISAANCCTQLLWMKHQLEDYQINANSIPIYCDNTA 1596
+ + S K +S+ EAE Y A+ T + +K E + N I + D+
Sbjct: 1668 IFNVYSNKSTNRCVSSTEAELHAIYEGYADSETLKVTLKELGEG---DNNDIVMITDSKP 1724
Query: 1597 AICLSKNPILHSRAKHIEIKHHFIRDYVQKGILDIQFIDTEHQWADIFTKPLSVERF 1653
AI + K IK I++ +++ + + I + AD+ TKP+S F
Sbjct: 1725 AIQGLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLKITGKGNIADLLTKPVSASDF 1781
Score = 81.6 bits (200), Expect = 2e-14
Identities = 142/609 (23%), Positives = 249/609 (40%), Gaps = 88/609 (14%)
Query: 404 ENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASKEELK 463
ENE+ KI + + L E L + T ++ + +D +K+Q + ++ S+ E
Sbjct: 189 ENEILCKI----IKEGLGESLDIMNTNTTDIF----RIIDGLKKQNIEVCMVEMSELEPG 240
Query: 464 GFNLISTTYEDRLKSLCQKLQE----------KCDKGSGNKHEIALDDFIMAGIDRSKVA 513
L+ TT + L KLQ+ KC + N + L + IM + S
Sbjct: 241 EKVLVDTTCRNSAL-LMNKLQKLVLMEKWIFSKCCQDCPNLKDY-LQEAIMGTLHESLRN 298
Query: 514 SMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTADQSKSEA-- 571
S+ Y N +G E E+ + + + + D + + DQ ++
Sbjct: 299 SVKQRLY-NIPHDVGIDHE---EFLINTVIETVID----------LSPIADDQIENSCMY 344
Query: 572 --SGSQAKITSKPENLKIKVMTKSDPKSQKITILK-RSETVHQNLIKPESKIPKQKDQKN 628
S I K + + +T+ P SQK I K + H + ++ E K + +++
Sbjct: 345 CKSVFHCSINCKKKPNRNLGLTR--PISQKPIIYKVHRDNNHLSPVQNEQKSWNKTQKRS 402
Query: 629 KAATASEKT--IPKGVKPKVLNDQKTLSIHPKVQGRKST----IG-NSSISINNVWLVDG 681
S+K I G + ND KTL +H +ST IG NSS+S+ +
Sbjct: 403 NKVYNSKKLVIIDTGSGVNITND-KTL-LHNYEDSNRSTRFFGIGKNSSVSVKGYGYIK- 459
Query: 682 LKHNLLSISQFCDNGYDVT------------FSKTNCTLVNK----DDKSITFKGKRVEN 725
+K+ + C Y V KT L K +K I K K V
Sbjct: 460 IKNGHNNTDNKCLLTYYVPEEESTIISCYDLAKKTKMVLSRKYTRLGNKIIKIKTKIVNG 519
Query: 726 VYKINFSDLA-----DQKVVCL-------LSMNDKKWVW---HKRLGHANWRLI-SKISK 769
V + ++L D K+ + +N + HKR+GH + I + I
Sbjct: 520 VIHVKMNELIERPSDDSKINAIKPTSSPGFKLNKRSITLEDAHKRMGHTGIQQIENSIKH 579
Query: 770 LQLVKGLPNIDYHSDALCGACQKGKIVKSSFKSKDIVSTS---RPLELLHIDLFGPVNTA 826
+ L I ++ C C+ K K + + + + S P +D+FGPV+++
Sbjct: 580 NHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTDHEPGSSWCMDIFGPVSSS 639
Query: 827 SLYGSKYGLVIVDDYSRW--TWVKFIKSKDYACEVFSSFCTQIQSEKELKILKVRSDHGG 884
+ +Y L++VD+ +R+ T F K+ + ++++ + K+ ++ SD G
Sbjct: 640 NADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQYVETQFDRKVREINSDRGT 699
Query: 885 EFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEA 944
EF N+ E + GI H +S + NG ER RT+ A T++ ++NL FW A
Sbjct: 700 EFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITDATTLLRQSNLRVKFWEYA 759
Query: 945 VNTSCYIQN 953
V ++ I+N
Sbjct: 760 VTSATNIRN 768
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 123 bits (309), Expect = 4e-27
Identities = 157/689 (22%), Positives = 292/689 (41%), Gaps = 75/689 (10%)
Query: 1025 QCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEIT 1084
Q E+ + R P TS SESN+ + P ++ + + + P ++
Sbjct: 652 QTSEQETEKRIIHRSPSIDTSS-SESNSLHHVVPIKTSDTCPKENTEESIIADLPLPDLP 710
Query: 1085 PE-----AESNSEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDS-- 1137
PE ++S E P Q S+ D+ K +E I S+D+
Sbjct: 711 PEPPTELSDSFKELPPINSRQTNSSLGGIGDSNAYTTINSKKRSLEDNETEIKVSRDTWN 770
Query: 1138 --------PRRTRSHFRQEESLIGLLSIIEPKTV---EEALS-------DDGWILAMQEE 1179
P R++ ++ + SI +T +EA++ + +I A +E
Sbjct: 771 TKNMRSLEPPRSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKE 830
Query: 1180 LNQFQRNDVWDLVPKPFQKNIIGTKWVFRNKL--NEQGEVTRNKARLVAQGYSQQEGIDY 1237
+NQ + WD K + + I K V + N + + T +KAR VA+G Q
Sbjct: 831 VNQLLKMKTWD-TDKYYDRKEIDPKRVINSMFIFNRKRDGT-HKARFVARGDIQHPDTYD 888
Query: 1238 TETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKHPN 1297
+ + A+ LS A+++ + Q+D+ SA+L I+EE+Y++ PP L +
Sbjct: 889 SGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP---HLGMND 945
Query: 1298 HVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVDDI 1357
+ +LKKSLYGLKQ+ WY+ + ++LIK + G + + + + + ++VDD+
Sbjct: 946 KLIRLKKSLYGLKQSGANWYETIKSYLIK---QCGMEEVRGWSCVFENSQVTICLFVDDM 1002
Query: 1358 IFGSTNASLCKEFSKLMQDEFEMSMMG------ELKF-FLGIQINQSKEGVYVHQTKYTK 1410
+ S N + K ++ +++ ++ E+++ LG++I + KY K
Sbjct: 1003 VLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEEIQYDILGLEIKYQR-------GKYMK 1055
Query: 1411 ELLKKFKLEDCKVMNTPMHP--TCTLSKEDTGTVVDQK--------------LYRGMIGS 1454
++ E +N P++P + G +DQ+ + +IG
Sbjct: 1056 LGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGL 1115
Query: 1455 LLYLTAS-RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDY--- 1510
Y+ R D+L+ + A+ P + L + +++ T + L++ KS
Sbjct: 1116 ASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPT 1175
Query: 1511 -KLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAANCCT 1569
KL+ DA Y G++ KS GN L +I S K + ST EAE + +
Sbjct: 1176 NKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVP 1234
Query: 1570 QLLWMKHQLEDYQINANSIPIYCD--NTAAICLSKNPILHSRAKHIEIKHHFIRDYVQKG 1627
L + + +++ + + D +T +I +S N R + K +RD V
Sbjct: 1235 LLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNE-EKFRNRFFGTKAMRLRDEVSGN 1293
Query: 1628 ILDIQFIDTEHQWADIFTKPLSVERFDFI 1656
L + +I+T+ AD+ TKPL ++ F +
Sbjct: 1294 HLHVCYIETKKNIADVMTKPLPIKTFKLL 1322
Score = 95.9 bits (237), Expect = 8e-19
Identities = 97/406 (23%), Positives = 170/406 (40%), Gaps = 33/406 (8%)
Query: 753 HKRLGHANWRLISKISKLQLVKGLPN--------IDYHS-DALCGACQKGKIVKSSFKSK 803
H+ L HAN + I K + IDY D L G K + +K S
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKY 231
Query: 804 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFS 861
++ P + LH D+FGPV+ Y + D+ +++ WV + + +D +VF+
Sbjct: 232 Q--NSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFT 289
Query: 862 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 921
+ I+++ + +L ++ D G E+ N F EK+GI +++ + +GV ER NR
Sbjct: 290 TILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNR 349
Query: 922 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 981
TL + RT + + L H W A+ S ++N + P +K+A + +IS
Sbjct: 350 TLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLP 408
Query: 982 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETQCVEESMHVKFDDREP 1040
FG I+N + K + G L S S Y +Y S + V+ + +V +E
Sbjct: 409 FG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKE- 466
Query: 1041 ESKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPEAEITPEAESNSEAEPSPKVQ 1100
+Q +A T D + + E++ S + I + + S+ E P
Sbjct: 467 --SRLDQFNYDALTFDEDLNRLTASYHSFIASNEIQQSNDLNIESDHDFQSDIELHP--- 521
Query: 1101 NESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSHFR 1146
E ++ + + P S H E DS R ++++ R
Sbjct: 522 -----EQLRNVLSKAVSPTDSTPPSTHTE------DSKRVSKTNIR 556
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 123 bits (309), Expect = 4e-27
Identities = 155/690 (22%), Positives = 293/690 (42%), Gaps = 86/690 (12%)
Query: 1034 KFDDREPESKTSEQSESNAGTTDSEDASESDQP-----SDSEKYTE---VESSPEAEITP 1085
+ D+E E + +S S + ++S + P + SE+ TE + P ++ P
Sbjct: 652 QISDQETEKRIIHRSPSIDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPP 711
Query: 1086 EAESN-----SEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDS--- 1137
E+ + E P Q S+ D+ K +E I S+D+
Sbjct: 712 ESPTEFPDPFKELPPINSHQTNSSLGGIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNT 771
Query: 1138 -------PRRTRSHFRQEESLIGLLSIIEPKTV---EEALS-------DDGWILAMQEEL 1180
P R++ ++ + SI +T +EA++ + +I A +E+
Sbjct: 772 KNMRSLEPPRSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEV 831
Query: 1181 NQFQRNDVWDLVPKPFQKNI-----IGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGI 1235
NQ + WD +K I I + ++F K + +KAR VA+G Q
Sbjct: 832 NQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDT 886
Query: 1236 DYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKH 1295
+ + A+ LS A+++ + Q+D+ SA+L I+EE+Y++ PP L
Sbjct: 887 YDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP---HLGM 943
Query: 1296 PNHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVD 1355
+ + +LKKSLYGLKQ+ WY+ + ++LIK + G + + K + + ++VD
Sbjct: 944 NDKLIRLKKSLYGLKQSGANWYETIKSYLIK---QCGMEEVRGWSCVFKNSQVTICLFVD 1000
Query: 1356 DIIFGSTNASLCKEFSKLMQDEFEMSMM------GELKF-FLGIQINQSKEGVYVHQTKY 1408
D+I S + + K+ ++ +++ ++ E+++ LG++I + KY
Sbjct: 1001 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQR-------GKY 1053
Query: 1409 TKELLKKFKLEDCKVMNTPMHP--TCTLSKEDTGTVVDQ------------KLY--RGMI 1452
K ++ E +N P++P + G +DQ K++ + +I
Sbjct: 1054 MKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLI 1113
Query: 1453 GSLLYLTAS-RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSL--- 1508
G Y+ R D+L+ + A+ P L + +++ T + L++ K+
Sbjct: 1114 GLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTE 1173
Query: 1509 -DYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAANC 1567
D KL+ DA Y G++ KS GN L +I S K + ST EAE + +
Sbjct: 1174 PDNKLVAISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISES 1232
Query: 1568 CTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHS-RAKHIEIKHHFIRDYVQK 1626
L + + +++ + D+ + I + K+ R + K +RD V
Sbjct: 1233 VPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSG 1292
Query: 1627 GILDIQFIDTEHQWADIFTKPLSVERFDFI 1656
L + +I+T+ AD+ TKPL ++ F +
Sbjct: 1293 NNLYVYYIETKKNIADVMTKPLPIKTFKLL 1322
Score = 95.5 bits (236), Expect = 1e-18
Identities = 90/388 (23%), Positives = 166/388 (42%), Gaps = 34/388 (8%)
Query: 753 HKRLGHANWRLISKISKLQLVKGLPN--------IDYHS-DALCGACQKGKIVKSSFKSK 803
H+ L HAN + I K + IDY D L G K + +K S
Sbjct: 172 HRMLAHANAQTIRYSLKNNTITYFNESDVDRSSAIDYQCPDCLIGKSTKHRHIKGSRLKY 231
Query: 804 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFS 861
++ P + LH D+FGPV+ Y + D+ +++ WV + + +D +VF+
Sbjct: 232 Q--NSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFT 289
Query: 862 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 921
+ I+++ + +L ++ D G E+ N F EK+GI +++ + +GV ER NR
Sbjct: 290 TILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNR 349
Query: 922 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 981
TL + RT + + L H W A+ S ++N + P +K+A + +IS
Sbjct: 350 TLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLP 408
Query: 982 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETQCVEESMHVKFDDREP 1040
FG I+N + K + G L S S Y +Y S + V+ + +V +E
Sbjct: 409 FG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKES 467
Query: 1041 E---------------SKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPE--AEI 1083
++ + +S + + + + + + SD + +++E PE +
Sbjct: 468 RLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNV 527
Query: 1084 TPEAESNSEAEPSPKVQNESASEDFQDN 1111
+A S +++ P P E + + N
Sbjct: 528 LSKAVSPTDSTP-PSTHTEDSKRVSKTN 554
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 121 bits (304), Expect = 1e-26
Identities = 155/690 (22%), Positives = 292/690 (41%), Gaps = 86/690 (12%)
Query: 1034 KFDDREPESKTSEQSESNAGTTDSEDASESDQP-----SDSEKYTE---VESSPEAEITP 1085
+ D+E E + +S S + ++S + P + SE+ TE + P ++ P
Sbjct: 1079 QISDQETEKRIIHRSPSIDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPP 1138
Query: 1086 EAESN-----SEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDS--- 1137
E+ + E P Q S+ D+ K +E I S+D+
Sbjct: 1139 ESPTEFPDPFKELPPINSHQTNSSLGGIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNT 1198
Query: 1138 -------PRRTRSHFRQEESLIGLLSIIEPKTV---EEALS-------DDGWILAMQEEL 1180
P R++ ++ + SI +T +EA++ + +I A +E+
Sbjct: 1199 KNMRSLEPPRSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEV 1258
Query: 1181 NQFQRNDVWDLVPKPFQKNI-----IGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGI 1235
NQ + WD +K I I + ++F K + +KAR VA+G Q
Sbjct: 1259 NQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDT 1313
Query: 1236 DYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKH 1295
T + A+ LS A+++ + Q+D+ SA+L I+EE+Y++ PP L
Sbjct: 1314 YDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP---HLGM 1370
Query: 1296 PNHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVD 1355
+ + +LKKS YGLKQ+ WY+ + ++LIK + G + + K + + ++VD
Sbjct: 1371 NDKLIRLKKSHYGLKQSGANWYETIKSYLIK---QCGMEEVRGWSCVFKNSQVTICLFVD 1427
Query: 1356 DIIFGSTNASLCKEFSKLMQDEFEMSMM------GELKF-FLGIQINQSKEGVYVHQTKY 1408
D+I S + + K+ ++ +++ ++ E+++ LG++I + KY
Sbjct: 1428 DMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQR-------GKY 1480
Query: 1409 TKELLKKFKLEDCKVMNTPMHP--TCTLSKEDTGTVVDQ------------KLY--RGMI 1452
K ++ E +N P++P + G +DQ K++ + +I
Sbjct: 1481 MKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLI 1540
Query: 1453 GSLLYLTAS-RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSL--- 1508
G Y+ R D+L+ + A+ P L + +++ T + L++ K+
Sbjct: 1541 GLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTE 1600
Query: 1509 -DYKLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAANC 1567
D KL+ DA Y G++ KS GN L +I S K + ST EAE + +
Sbjct: 1601 PDNKLVAISDASY-GNQPYYKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISES 1659
Query: 1568 CTQLLWMKHQLEDYQINANSIPIYCDNTAAICLSKNPILHS-RAKHIEIKHHFIRDYVQK 1626
L + + +++ + D+ + I + K+ R + K +RD V
Sbjct: 1660 VPLLNNLSYLIQELNKKPIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSG 1719
Query: 1627 GILDIQFIDTEHQWADIFTKPLSVERFDFI 1656
L + +I+T+ AD+ TKPL ++ F +
Sbjct: 1720 NNLYVYYIETKKNIADVMTKPLPIKTFKLL 1749
Score = 96.7 bits (239), Expect = 5e-19
Identities = 90/388 (23%), Positives = 168/388 (43%), Gaps = 34/388 (8%)
Query: 753 HKRLGHANWRLISKISKLQLVKGLPN--------IDYHS-DALCGACQKGKIVKSSFKSK 803
H+ L HAN + I K + IDY D L G K + +K S
Sbjct: 599 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKY 658
Query: 804 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFS 861
++ P + LH D+FGPV+ Y + D+ +++ WV + + +D +VF+
Sbjct: 659 Q--NSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFT 716
Query: 862 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 921
+ I+++ + +L ++ D G E+ N F EK+GI +++ + +GV ER NR
Sbjct: 717 TILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNR 776
Query: 922 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 981
TL + RT + + L H W A+ S ++N + P +K+A + +IS
Sbjct: 777 TLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLP 835
Query: 982 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETQCVEESMHVKFDDREP 1040
FG I+N + K + G L S S Y +Y S + V+ + +V +E
Sbjct: 836 FG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKES 894
Query: 1041 E---------------SKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPE--AEI 1083
++ + +S + + +++++ + SD + +++E PE +
Sbjct: 895 RLDQFNYDALTFDEDLNRLTASYQSFIASNEIQESNDLNIESDHDFQSDIELHPEQPRNV 954
Query: 1084 TPEAESNSEAEPSPKVQNESASEDFQDN 1111
+A S +++ P P E + + N
Sbjct: 955 LSKAVSPTDSTP-PSTHTEDSKRVSKTN 981
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 121 bits (303), Expect = 2e-26
Identities = 155/691 (22%), Positives = 293/691 (41%), Gaps = 88/691 (12%)
Query: 1034 KFDDREPESKTSEQSESNAGTTDSEDASESDQP-----SDSEKYTE---VESSPEAEITP 1085
+ D+E E + +S S + ++S + P + SE+ TE + P ++ P
Sbjct: 1079 QISDQETEKRIIHRSPSIDASPPENNSSHNIVPIKTPTTVSEQNTEESIIADLPLPDLPP 1138
Query: 1086 EAESN-----SEAEPSPKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDS--- 1137
E+ + E P Q S+ D+ K +E I S+D+
Sbjct: 1139 ESPTEFPDPFKELPPINSRQTNSSLGGIGDSNAYTTINSKKRSLEDNETEIKVSRDTWNT 1198
Query: 1138 -------PRRTRSHFRQEESLIGLLSIIEPKTV---EEALS-------DDGWILAMQEEL 1180
P R++ ++ + SI +T +EA++ + +I A +E+
Sbjct: 1199 KNMRSLEPPRSKKRIHLIAAVKAVKSIKPIRTTLRYDEAITYNKDIKEKEKYIEAYHKEV 1258
Query: 1181 NQFQRNDVWDLVPKPFQKNI-----IGTKWVFRNKLNEQGEVTRNKARLVAQGYSQQEGI 1235
NQ + WD +K I I + ++F K + +KAR VA+G Q
Sbjct: 1259 NQLLKMKTWDTDEYYDRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDT 1313
Query: 1236 DYTETFAPVARLEAIRLLLSYAINHGIILYQMDVKSAFLNGVIEEEVYVKQPPGFEDLKH 1295
+ + A+ LS A+++ + Q+D+ SA+L I+EE+Y++ PP L
Sbjct: 1314 YDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPP---HLGM 1370
Query: 1296 PNHVYKLKKSLYGLKQAPRAWYDRLSNFLIKNDFERGQVDTTLFRRTLKKDILIVQIYVD 1355
+ + +LKKSLYGLKQ+ WY+ + ++LI+ + G + + K + + ++VD
Sbjct: 1371 NDKLIRLKKSLYGLKQSGANWYETIKSYLIQ---QCGMEEVRGWSCVFKNSQVTICLFVD 1427
Query: 1356 DIIFGSTNASLCKEFSKLMQDEFEMSMMG------ELKF-FLGIQINQSKEGVYVHQTKY 1408
D++ S N + K + ++ +++ ++ E+++ LG++I + KY
Sbjct: 1428 DMVLFSKNLNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQR-------GKY 1480
Query: 1409 TKELLKKFKLEDCKVMNTPMHP--TCTLSKEDTGTVVDQK--------------LYRGMI 1452
K ++ E +N P++P + G +DQ+ + +I
Sbjct: 1481 MKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLI 1540
Query: 1453 GSLLYLTAS-RPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDY- 1510
G Y+ R D+L+ + A+ P + L + +++ T + L++ KS
Sbjct: 1541 GLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVK 1600
Query: 1511 ---KLIGFCDADYAGDRIERKSTSGNCQFLGENLISWASKKQATIAMSTAEAEYISAANC 1567
KL+ DA Y G++ KS GN L +I S K + ST EAE + +
Sbjct: 1601 PTNKLVVISDASY-GNQPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISES 1659
Query: 1568 CTQLLWMKHQLEDYQINANSIPIYCD--NTAAICLSKNPILHSRAKHIEIKHHFIRDYVQ 1625
L + + +++ + + D +T +I +S N R + K +RD V
Sbjct: 1660 VPLLNNLSYLIQELDKKPITKGLLTDSKSTISIIISNNE-EKFRNRFFGTKAMRLRDEVS 1718
Query: 1626 KGILDIQFIDTEHQWADIFTKPLSVERFDFI 1656
L + +I+T+ AD+ TKPL ++ F +
Sbjct: 1719 GNHLHVCYIETKKNIADVMTKPLPIKTFKLL 1749
Score = 95.5 bits (236), Expect = 1e-18
Identities = 90/388 (23%), Positives = 166/388 (42%), Gaps = 34/388 (8%)
Query: 753 HKRLGHANWRLISKISKLQLVKGLPN--------IDYHS-DALCGACQKGKIVKSSFKSK 803
H+ L HAN + I K + IDY D L G K + +K S
Sbjct: 599 HRMLAHANAQTIRYSLKNNTITYFNESDVDWSSAIDYQCPDCLIGKSTKHRHIKGSRLKY 658
Query: 804 DIVSTSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSRWTWVKFI--KSKDYACEVFS 861
++ P + LH D+FGPV+ Y + D+ +++ WV + + +D +VF+
Sbjct: 659 Q--NSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFT 716
Query: 862 SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILHEFSSPRTPQQNGVVERKNR 921
+ I+++ + +L ++ D G E+ N F EK+GI +++ + +GV ER NR
Sbjct: 717 TILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNR 776
Query: 922 TLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLEKTAYELFKGRRPNISYFHQ 981
TL + RT + + L H W A+ S ++N + P +K+A + +IS
Sbjct: 777 TLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSL-ASPKSKKSARQHAGLAGLDISTLLP 835
Query: 982 FGCTCYILNTKDYLKKFDAKAQRGIFLGYSERSKAYRVY-NSETQCVEESMHVKFDDREP 1040
FG I+N + K + G L S S Y +Y S + V+ + +V +E
Sbjct: 836 FG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKES 894
Query: 1041 E---------------SKTSEQSESNAGTTDSEDASESDQPSDSEKYTEVESSPE--AEI 1083
++ + +S + + + + + + SD + +++E PE +
Sbjct: 895 RLDQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNV 954
Query: 1084 TPEAESNSEAEPSPKVQNESASEDFQDN 1111
+A S +++ P P E + + N
Sbjct: 955 LSKAVSPTDSTP-PSTHTEDSKRVSKTN 981
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 110 bits (275), Expect = 3e-23
Identities = 53/99 (53%), Positives = 68/99 (68%)
Query: 1159 EPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNIIGTKWVFRNKLNEQGEVT 1218
EPK+V AL D GW AMQEEL+ RN W LVP P +NI+G KWVF+ KL+ G +
Sbjct: 27 EPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDGTLD 86
Query: 1219 RNKARLVAQGYSQQEGIDYTETFAPVARLEAIRLLLSYA 1257
R KARLVA+G+ Q+EGI + ET++PV R IR +L+ A
Sbjct: 87 RLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVA 125
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 63.2 bits (152), Expect = 6e-09
Identities = 30/82 (36%), Positives = 48/82 (57%)
Query: 1456 LYLTASRPDILFSVCLCARFQSDPRESHLTAVKRIFRYLKGTTNLGLLYRKSLDYKLIGF 1515
+YLT +RPD+ F+V ++F S R + + AV ++ Y+KGT GL Y + D +L F
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 1516 CDADYAGDRIERKSTSGNCQFL 1537
D+D+A R+S +G C +
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLV 82
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 60.1 bits (144), Expect = 5e-08
Identities = 53/199 (26%), Positives = 87/199 (43%), Gaps = 16/199 (8%)
Query: 788 GACQKGKIVKSSFKSKDIV----STSRPLELLHIDLFGPVNTASLYGSKYGLVIVDDYSR 843
G CQ+ I +S K+ + +P + ID GP+ + G Y LV+VD +
Sbjct: 648 GRCQQCLITNASNKASGPILRPDRPQKPFDKFFIDYIGPLPPSQ--GYLYVLVVVDGMTG 705
Query: 844 WTWVKFIKSKDYACEVFS-SFCTQIQSEKELKILKVRSDHGGEFENEPFELFCEKHGILH 902
+TW+ K+ + V S + T I K + SD G F + F + ++ GI
Sbjct: 706 FTWLYPTKAPSTSATVKSLNVLTSIAIPKV-----IHSDQGAAFTSSTFAEWAKERGIHL 760
Query: 903 EFSSPRTPQQNGVVERKNRTLQEMARTMIHENNLAKHFWAEAVNTSCYIQNRIYIRPMLE 962
EFS+P PQ VERKN ++ + ++ W + + N Y P+L+
Sbjct: 761 EFSTPYHPQSGSKVERKNSDIKRLLTKLLVGRPTK---WYDLLPVVQLALNNTY-SPVLK 816
Query: 963 KTAYELFKGRRPNISYFHQ 981
T ++L G N + +Q
Sbjct: 817 YTPHQLLFGIDSNTPFANQ 835
>MST2_DROHY (Q08696) Axoneme-associated protein mst101(2)
Length = 1391
Score = 60.1 bits (144), Expect = 5e-08
Identities = 97/472 (20%), Positives = 168/472 (35%), Gaps = 22/472 (4%)
Query: 185 DHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIAL 244
+ ++K ++ + + K A +E + E+L +K ET+KK K +A
Sbjct: 432 EELAKNIKKAAEKKKCKEAAKKEKEAAERKKCEELAKKIKKAAEKKKCEETAKKGKEVAE 491
Query: 245 PSK--------GKTSKSSKAYKASESEEESPDGDSDEDQSVKM--AMLSNKLEYLARKQK 294
K K K K ++ E+E+ + E + K A K E A+K+K
Sbjct: 492 RKKCEELAKKIKKAEIKKKCKKLAKKEKETAEKKKCEKAAKKRKEAAEKKKCEKAAKKRK 551
Query: 295 KFLSKRGSYKNSKKEDQKG-CFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQI 353
+ K+ K++KK + C+K + + +K + K K K +
Sbjct: 552 EAAEKKKCEKSAKKRKEAAEKKKCEKAAKERKEAAEKKKCEEAAKKEKEVAERKKCEELA 611
Query: 354 KKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPR 413
KK A + E+ ++EA + K + + + ++ E E K
Sbjct: 612 KKIKKAAEKKKCKEAAKKEKEAAEREKCGELAKKIKKAAEKKKCKKLAKKEKETAEKKKC 671
Query: 414 QELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYE 473
++ KE E + KEK K+ + + K + E K L +
Sbjct: 672 EKAAKKRKEAA---EKKKCAEAAKKEKEAAEKKKCEEAAKKEKEAAERKKCEELAKKIKK 728
Query: 474 DRLKSLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEK 533
K C+KL +K G NK + G + K A + K ++K
Sbjct: 729 AAEKKKCKKLAKKKKAGEKNK--------LKKGNKKGKKALKEKKKCRELAKKKAAEKKK 780
Query: 534 SKEYSLKSYCDCIKDGLKSTFVPEGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTKS 593
KE + K K + T A K+ +A K E K ++
Sbjct: 781 CKEAAKKEKEAAEKKKCEKTAKKRKEEAEKKKCEKTAKKRKEAAEKKKCEKAAKKRKEEA 840
Query: 594 DPKSQKITILKRSETVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPK 645
+ K + T KR ET + + +K KQ +K K A++K K K
Sbjct: 841 EKKKCEKTAKKRKETAEKKKCEKAAKKRKQAAEKKKCEKAAKKRKEAAEKKK 892
Score = 55.5 bits (132), Expect = 1e-06
Identities = 90/440 (20%), Positives = 170/440 (38%), Gaps = 48/440 (10%)
Query: 224 KVHEMSLNEHETSKKSKSIALPSK-GKTSKSSKAYKASESEEESPDGDSDED--QSVKMA 280
K E + E E +++ K L K K ++ K +A++ E+E+ + + + + +K A
Sbjct: 590 KCEEAAKKEKEVAERKKCEELAKKIKKAAEKKKCKEAAKKEKEAAEREKCGELAKKIKKA 649
Query: 281 MLSNKLEYLARKQKKFLSKRGSYKNSKKE----DQKGCFNCKKPGHFIVDCPDLQKEKFK 336
K + LA+K+K+ K+ K +KK ++K C K + ++ K
Sbjct: 650 AEKKKCKKLAKKEKETAEKKKCEKAAKKRKEAAEKKKCAEAAKKEKEAAEKKKCEEAAKK 709
Query: 337 GKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSE 396
K + K+IKK+ E + + K++A + K G ++ +
Sbjct: 710 EKEAAERKKCEELAKKIKKAA----EKKKCKKLAKKKKAGEKNKLKKG------NKKGKK 759
Query: 397 AESDSEDENEVYSK--IPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLE 454
A + + E+ K +++ ++ K+ E + E T K K K+ + T +
Sbjct: 760 ALKEKKKCRELAKKKAAEKKKCKEAAKKEKEAAEKKKCEKTAKKRKEEAEKKKCEKTAKK 819
Query: 455 LKASKEELKGFNLISTTYEDRLKSLCQKL---------QEKCDKGSGNKHEIALDDFIMA 505
K + E+ K E+ K C+K ++KC+K + + + A
Sbjct: 820 RKEAAEKKKCEKAAKKRKEEAEKKKCEKTAKKRKETAEKKKCEKAAKKRKQAAEKKKCEK 879
Query: 506 GIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTAD 565
+ K A+ + K + +K KE + K C+ K A+
Sbjct: 880 AAKKRKEAA--------EKKKCAEAAKKEKELAEKKKCEEAAKKEKE----------VAE 921
Query: 566 QSKSEASGSQAKITSKPENLKIKVMTKSDPKSQKITILKRSETVHQNLIKPESKIPKQKD 625
+ K E KI E K K + K + K+ + LK+ + K K K+
Sbjct: 922 RKKCEELAK--KIKKAAEKKKCKKLAKKEKKAGEKNKLKKKAGKGKKKCKKLGKKSKRAA 979
Query: 626 QKNKAATASEKTIPKGVKPK 645
+K K A A++K K K
Sbjct: 980 EKKKCAEAAKKEKEAATKKK 999
Score = 50.8 bits (120), Expect = 3e-05
Identities = 91/426 (21%), Positives = 155/426 (36%), Gaps = 50/426 (11%)
Query: 224 KVHEMSLNEHETSKKSKSIALPSKGKT-SKSSKAYKASESEEESPDGDSDED--QSVKMA 280
K + + E ++K K K K ++ K +A++ E+E + E+ + +K A
Sbjct: 876 KCEKAAKKRKEAAEKKKCAEAAKKEKELAEKKKCEEAAKKEKEVAERKKCEELAKKIKKA 935
Query: 281 MLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSM 340
K + LA+K+KK G KK+ KG CKK G + +K K
Sbjct: 936 AEKKKCKKLAKKEKK----AGEKNKLKKKAGKGKKKCKKLGKKSKRAAEKKKCAEAAKKE 991
Query: 341 KSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESD 400
K + K ++ KK A + E +EA + + + + E
Sbjct: 992 KEAATKKKCEERAKKQKEAAEKKQCEERAKKLKEAAEQKQCEERAKKLKEAAEKKQCEER 1051
Query: 401 SEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASKE 460
++ E + +E LKE E + E KEK KQ + +LK + E
Sbjct: 1052 AKKLKEAAEQKQCEERAKKLKEAA---EKKQCEERAKKEKEAAEKKQCEERAKKLKEAAE 1108
Query: 461 ELKGFNLISTTYEDRLKSLCQKLQEK-CDKGSGNKHEIALDDFIMAGIDRSKVASMIYST 519
+ + E+R K + ++K C++ + + E A + K A T
Sbjct: 1109 KKQ--------CEERAKKEKEAAEKKRCEEAAKREKEAAEKKKCAEAAKKEKEA-----T 1155
Query: 520 YKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTADQSKSEASGSQAKIT 579
K K + +K KE + K C A+ +K E +Q K
Sbjct: 1156 EKQK---CAEAAKKEKEAAEKKKC--------------------AEAAKREKEAAQKK-- 1190
Query: 580 SKPENLKIKVMTKSDPKSQKITILKRSETVHQNLIKPESKIPKQKDQKNKAATASEKTIP 639
K +L K ++ K + K E + +K K+ +K K A A++K
Sbjct: 1191 -KCADLAKKEQEPAEMKKCEEAAKKEKEAAEKQKCAKAAKKEKEAAEKKKCAEAAKKEQE 1249
Query: 640 KGVKPK 645
K K
Sbjct: 1250 AAEKKK 1255
Score = 50.4 bits (119), Expect = 4e-05
Identities = 98/459 (21%), Positives = 167/459 (36%), Gaps = 76/459 (16%)
Query: 201 KVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKAS 260
K A EEAK + +V +K E KK K + K + K A A
Sbjct: 306 KEQAEEEAK------IRGVVKEVK---KKCKEKALKKKCKDLGRKMKEEAEKKKCAALAK 356
Query: 261 ESEEESPDGDSDE-DQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKK 319
+ +EE E + K A K E A K+KK K+ K +K ++K KK
Sbjct: 357 KQKEEDEKKACKELAKKKKEADEKKKCEEAANKEKKAAEKKKCEKAAK--ERKEAAEKKK 414
Query: 320 PGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDA 379
C + K++ + K + K IKK+ E + + KE+ +
Sbjct: 415 -------CEEAAKKEKEAAERK---KCEELAKNIKKAA----EKKKCKEAAKKEKEAAER 460
Query: 380 KAAVGLVATVSSEA-VSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLK 438
K L + A + E ++ EV + +EL +K+ ++K
Sbjct: 461 KKCEELAKKIKKAAEKKKCEETAKKGKEVAERKKCEELAKKIKK------------AEIK 508
Query: 439 EKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKL---------QEKCDK 489
+K L K++K T + K K K E K C+K ++KC+K
Sbjct: 509 KKCKKLAKKEKETAEKKKCEKAAKK-------RKEAAEKKKCEKAAKKRKEAAEKKKCEK 561
Query: 490 GSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDG 549
+ + E A K A+ + K + +K KE + + C+ +
Sbjct: 562 SAKKRKEAAEKKKCEKAAKERKEAA--------EKKKCEEAAKKEKEVAERKKCEELAKK 613
Query: 550 LKSTFVPEGTNAVTADQSKSEASGSQAKITSKPE---NLKIKVMTKSDPKSQKITILKRS 606
+K A++ K + + + K ++ E L K+ ++ K K K
Sbjct: 614 IKK----------AAEKKKCKEAAKKEKEAAEREKCGELAKKIKKAAEKKKCKKLAKKEK 663
Query: 607 ETVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPK 645
ET + + +K K+ +K K A A++K K K
Sbjct: 664 ETAEKKKCEKAAKKRKEAAEKKKCAEAAKKEKEAAEKKK 702
Score = 41.2 bits (95), Expect = 0.024
Identities = 82/421 (19%), Positives = 159/421 (37%), Gaps = 44/421 (10%)
Query: 224 KVHEMSLNEHETSKKSKSIALPSKGK-TSKSSKAYKASESEEESPDGDSDED--QSVKMA 280
K + + E ++K K K K ++ K +A++ E+E+ + E+ + +K A
Sbjct: 670 KCEKAAKKRKEAAEKKKCAEAAKKEKEAAEKKKCEEAAKKEKEAAERKKCEELAKKIKKA 729
Query: 281 MLSNKLEYLARKQK---KFLSKRGSYKNSK--KEDQKGCFNCKKPGHFIVDCPDLQKEKF 335
K + LA+K+K K K+G+ K K KE +K KK C + K++
Sbjct: 730 AEKKKCKKLAKKKKAGEKNKLKKGNKKGKKALKEKKKCRELAKKKAAEKKKCKEAAKKEK 789
Query: 336 KGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVS 395
+ K ++K RK+ + + ++K++ + AK E +
Sbjct: 790 EAAEKKKCEKTAKKRKEEAEKKKCEKTAKKRKEAAEKKKCEKAAKKRKEEAEKKKCEKTA 849
Query: 396 EAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLEL 455
+ ++ ++ + +++ K+ + R K+K + K++K L E
Sbjct: 850 KKRKETAEKKKCEKAAKKRKQAAEKKKCEKAAKKRKEAAE--KKKCAEAAKKEKE-LAEK 906
Query: 456 KASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRSKVASM 515
K +E K ++ + C++L +K K + K K+A
Sbjct: 907 KKCEEAAKKEKEVAE------RKKCEELAKKIKKAAEKK-------------KCKKLAK- 946
Query: 516 IYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTADQSKSEASGSQ 575
K K G + K K+ + K C K G KS E A + + EA+ +
Sbjct: 947 -----KEKKAG---EKNKLKKKAGKGKKKCKKLGKKSKRAAEKKKCAEAAKKEKEAATKK 998
Query: 576 AKITSKPENLKIKVMTKSDPKSQKITILKRSETVHQNLIKPESKIPKQKDQKNKAATASE 635
K E K ++ K + K E Q + +K K+ +K + ++
Sbjct: 999 -----KCEERAKKQKEAAEKKQCEERAKKLKEAAEQKQCEERAKKLKEAAEKKQCEERAK 1053
Query: 636 K 636
K
Sbjct: 1054 K 1054
Score = 38.1 bits (87), Expect = 0.20
Identities = 60/317 (18%), Positives = 124/317 (38%), Gaps = 31/317 (9%)
Query: 201 KVTAIEEAKDLNTLS--VEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYK 258
K + E K L+ ++ K +++ E + +K+K KGK K
Sbjct: 915 KEKEVAERKKCEELAKKIKKAAEKKKCKKLAKKEKKAGEKNKLKKKAGKGKKKCKKLGKK 974
Query: 259 ASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKK----EDQKGC 314
+ + E+ ++ + + K A K E A+KQK+ K+ + +KK +QK
Sbjct: 975 SKRAAEKKKCAEAAKKE--KEAATKKKCEERAKKQKEAAEKKQCEERAKKLKEAAEQK-- 1030
Query: 315 FNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 374
C++ + + + ++ + + K +K + + ++ KK A + E ++E
Sbjct: 1031 -QCEERAKKLKEAAEKKQCEERAKKLKEAAEQKQCEERAKKLKEAAEKKQCEERAKKEKE 1089
Query: 375 ADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFE-----H 429
A + + + + E ++ E E K +E KE +
Sbjct: 1090 AAEKKQCEERAKKLKEAAEKKQCEERAKKEKEAAEKKRCEEAAKREKEAAEKKKCAEAAK 1149
Query: 430 RTNELTDLKEKYVDLMKQQKSTLLELK---ASKEELKGFNLISTTYEDRLKSLCQKLQE- 485
+ E T+ K+K + K++K + K A+K E + + + L +K QE
Sbjct: 1150 KEKEATE-KQKCAEAAKKEKEAAEKKKCAEAAKREKE------AAQKKKCADLAKKEQEP 1202
Query: 486 ----KCDKGSGNKHEIA 498
KC++ + + E A
Sbjct: 1203 AEMKKCEEAAKKEKEAA 1219
>GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member 4
(Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (72.1
protein)
Length = 2230
Score = 60.1 bits (144), Expect = 5e-08
Identities = 118/568 (20%), Positives = 233/568 (40%), Gaps = 64/568 (11%)
Query: 138 KEAKALMLVHQYELFRMKDDES-IEEMYSRFQTLVSGLQILKKS----YVASDHVSKILR 192
+E L + Q ++++ E+ I+ M + ++LV+ + L+K A+ +
Sbjct: 1283 EEQNTLNISFQQATHQLEEKENQIKSMKADIESLVTEKEALQKEGGNQQQAASEKESCIT 1342
Query: 193 SLPSRWRPKVTAIEEAKD-LNTLSVEDLVSSLKVHEMSLN---EHETSKKSKSIALPSKG 248
L + A+ K+ L VE +SSL LN ++ S K A+ S
Sbjct: 1343 QLKKELSENINAVTLMKEELKEKKVE--ISSLSKQLTDLNVQLQNSISLSEKEAAISSLR 1400
Query: 249 KTSKSSKAYKASESEEESPDGDSDEDQSVK-MAMLSNKLEYLARKQKKFLSKRGSYKNSK 307
K K + ++ S D+ + + + + + + +KK S+ ++N+
Sbjct: 1401 KQYDEEKCELLDQVQDLSFKVDTLSKEKISALEQVDDWSNKFSEWKKKAQSRFTQHQNTV 1460
Query: 308 KEDQ-------KGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMAT 360
KE Q K + + + + + D Q ++F +K K + + K+S + T
Sbjct: 1461 KELQIQLELKSKEAYEKDEQINLLKEELDQQNKRFD--CLKGEMEDDKSKMEKKESNLET 1518
Query: 361 WEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSL 420
+L S++ E D + T+ E+++E + + ++ K ELV L
Sbjct: 1519 --ELKSQTARIMELEDHITQK------TIEIESLNEVLKNYNQQKDIEHK----ELVQKL 1566
Query: 421 KELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLC 480
+ L E + N + + +EK + L Q S EL+ K+EL+ NL + E+ LK+L
Sbjct: 1567 QHFQELGEEKDNRVKEAEEKILTLENQVYSMKAELETKKKELEHVNLSVKSKEEELKALE 1626
Query: 481 QKLQ-EKCDKGSGNKHEIALDDFIMAGIDRSKVASMIYSTYK-NKGKGIGYSEEKSKEYS 538
+L+ E K + K + + +A I + ++ M + KG SE +K
Sbjct: 1627 DRLESESAAKLAELKRKA---EQKIAAIKKQLLSQMEEKEEQYKKGTESHLSELNTKLQE 1683
Query: 539 LKSYCDCIKDGLKST--------FVPEGTNAVTA--DQSKSEASGSQAKITSKPENLKIK 588
+ +++ LKS VP V A +Q ++++ G K + ++ +
Sbjct: 1684 REREVHILEEKLKSVESSQSETLIVPRSAKNVAAYTEQEEADSQGCVQKTYEEKISVLQR 1743
Query: 589 VMTKSDPKSQKITILKRSETV----------HQNLIKPESKIPKQKDQKNKAATASEKTI 638
+T+ + Q++ ++ ETV + LIK E KQ + ++ E+
Sbjct: 1744 NLTEKEKLLQRVG-QEKEETVSSHFEMRCQYQERLIKLEHAEAKQHEDQSMIGHLQEELE 1802
Query: 639 PKGVKPKVLNDQKTLSIHPKVQGRKSTI 666
K K ++ Q H + +G K+ I
Sbjct: 1803 EKNKKYSLIVAQ-----HVEKEGGKNNI 1825
Score = 58.9 bits (141), Expect = 1e-07
Identities = 94/460 (20%), Positives = 197/460 (42%), Gaps = 46/460 (10%)
Query: 55 LDEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMKM 114
L+EEL +L+D D + + E +++ H Q K H++ SI RTE
Sbjct: 703 LEEEL-SVLKDQTDKMKQELEAKMDEQKNHHQQQVDSIIKEHEV------SIQRTEKALK 755
Query: 115 SDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSRFQTLVSGL 174
+ + + E K +KE HQ + ++ D I+ Q + L
Sbjct: 756 DQINQLELLLK------ERDKHLKE-------HQAHVENLEAD--IKRSEGELQQASAKL 800
Query: 175 QILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHE 234
+ + A+ +K ++ + K+ +E + L T V ++ + K L+ H+
Sbjct: 801 DVFQSYQSATHEQTKAYEEQLAQLQQKLLDLETERILLTKQVAEVEAQKKDVCTELDAHK 860
Query: 235 TSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQK 294
+ L + + E + DG+ +++Q+ ++ + + R+ +
Sbjct: 861 IQVQDLMQQLEKQNSEMEQKVKSLTQVYESKLEDGNKEQEQTKQILVEKENMILQMREGQ 920
Query: 295 KFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQ-- 352
K + + K S KED N + F ++K K K K M+ + ++
Sbjct: 921 KKEIEILTQKLSAKEDSIHILNEEYETKFKNQEKKMEKVKQKAKEMQETLKKKLLDQEAK 980
Query: 353 IKKSLMATWEDLDSESGSDKEEADDDAKA-AVGLVATVSSEAVSEAESDSEDENEVYSKI 411
+KK L T +L + + + A+A + G+ S+AVS E++ +++ E +++
Sbjct: 981 LKKELENTALELSQKEKQFNAKMLEMAQANSAGI-----SDAVSRLETNQKEQIESLTEV 1035
Query: 412 PRQELVDSLKELLSLFEHRTN-ELTDLKEKYVDLMKQQKSTLLELK-------ASKEELK 463
R+EL D ++S++E + N + +L+E + +++++ + ELK KEE+
Sbjct: 1036 HRRELND----VISIWEKKLNQQAEELQEIHEIQLQEKEQEVAELKQKILLFGCEKEEMN 1091
Query: 464 GFNLISTTYEDRLK--SLCQKLQEKCDKGSGNKHEIALDD 501
I+ E+ +K + +LQE+ + S + + +A D+
Sbjct: 1092 --KEITWLKEEGVKQDTTLNELQEQLKQKSAHVNSLAQDE 1129
Score = 35.8 bits (81), Expect = 1.0
Identities = 70/386 (18%), Positives = 150/386 (38%), Gaps = 50/386 (12%)
Query: 149 YELFRMKDDESIEEMYSRFQTLVS---------------GLQILKKSYVASDHVSKILRS 193
+E MK++E I ++ SR + + + + L+K+ + + R
Sbjct: 374 HETLEMKEEE-IAQLRSRIKQMTTQGEELREQKEKSERAAFEELEKALSTAQKTEEARRK 432
Query: 194 LPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKS 253
L + ++ IE+ + +S++ +S +K + + KKS + K +
Sbjct: 433 LKAEMDEQIKTIEKTSEEERISLQQELSRVKQEVV-----DVMKKSSEEQIAKLQKLHEK 487
Query: 254 SKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKG 313
A K E ++ + + + +K+A+ ++ EYL Q+K ++ S + E QK
Sbjct: 488 ELARKEQELTKKLQTREREFQEQMKVALEKSQSEYLKISQEK--EQQESLALEELELQKK 545
Query: 314 CFNCKKPGHFIVDCPDLQKE---------KFKGKSMKSSFNSSKFRKQIKKSLMATWEDL 364
+ DLQ+E + + KS + K + L A
Sbjct: 546 AILTESENKL----RDLQQEAETYRTRILELESSLEKSLQENKNQSKDLAVHLEAEKNKH 601
Query: 365 DSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELL 424
+ E E+ + ++ + +E + + + E E + QE LK+
Sbjct: 602 NKEITVMVEKHKTELESLKHQQDALWTEKLQVLKQQYQTEMEKLREKCEQEKETLLKDKE 661
Query: 425 SLFE-------HRTNELTDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLK 477
+F+ +T E D+K+ ++ + + S +L+ + EE + +D+
Sbjct: 662 IIFQAHIEEMNEKTLEKLDVKQTELESLSSELSEVLKARHKLEE------ELSVLKDQTD 715
Query: 478 SLCQKLQEKCDKGSGNKHEIALDDFI 503
+ Q+L+ K D+ N H+ +D I
Sbjct: 716 KMKQELEAKMDE-QKNHHQQQVDSII 740
>USO1_YEAST (P25386) Intracellular protein transport protein USO1
Length = 1790
Score = 59.3 bits (142), Expect = 9e-08
Identities = 129/681 (18%), Positives = 277/681 (39%), Gaps = 100/681 (14%)
Query: 6 ESVTTKYTSVKHDYDTADKKTDSGKAPKFNGDPEEFSWWKTNMYSFIMGLDE--ELWDIL 63
E++T K ++ +++ D+K + + E FS +T + + LDE +L D+L
Sbjct: 758 ENLTEKLIALTNEHKELDEKYQILNS-SHSSLKENFSILETELKNVRDSLDEMTQLRDVL 816
Query: 64 EDGVDDLDLDEEGAAIDRRIHTPAQKKLYKKHHKIRGIIVASIPRTEYMKMSDKSTAKAM 123
E D + + A ++ + Q+ K K I++ + E A+
Sbjct: 817 ETK----DKENQTALLEYKSTIHKQEDSIKTLEKGLETILSQKKKAEDGINKMGKDLFAL 872
Query: 124 FASLCANFEGSKKVKEAKALMLV-HQYELFRMKDDESIEEMYSRFQTLVSGLQILK---- 178
+ A E K +++ K V HQ E +K+D I + + + L+ +K
Sbjct: 873 SREMQAVEENCKNLQKEKDKSNVNHQKETKSLKED--IAAKITEIKAINENLEEMKIQCN 930
Query: 179 KSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKK 238
+H+SK L SR++ S ++LV+ L ++K
Sbjct: 931 NLSKEKEHISKELVEYKSRFQ---------------SHDNLVAKL------------TEK 963
Query: 239 SKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLS 298
KS+A K +++ KA E +S + S++++ L NK++ ++++++ F
Sbjct: 964 LKSLANNYKDMQAENESLIKAVE--------ESKNESSIQLSNLQNKIDSMSQEKENFQI 1015
Query: 299 KRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLM 358
+RGS + + ++ +K + ++ I+ D K++++ + S +++++ +
Sbjct: 1016 ERGSIEKNIEQLKKTISDLEQTKEEIISKSDSSKDEYESQ-------ISLLKEKLETATT 1068
Query: 359 ATWEDLD--SESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQEL 416
A E+++ SE +EE + + A L +NE+ +K+ E
Sbjct: 1069 ANDENVNKISELTKTREELEAELAAYKNL------------------KNELETKLETSE- 1109
Query: 417 VDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRL 476
+LKE+ EH LKE+ + L K+ T +L + + L+ + +ED
Sbjct: 1110 -KALKEVKENEEH-------LKEEKIQLEKEATETKQQLNSLRANLES---LEKEHEDLA 1158
Query: 477 KSLCQKLQEKCDKGSGNKHEIA-LDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSK 535
L + ++ +K EI+ L+D I + ++ + + K + + E+
Sbjct: 1159 AQLKKYEEQIANKERQYNEEISQLNDEITSTQQENESIKKKNDELEGEVKAMKSTSEEQS 1218
Query: 536 EYSLKSYCDCIKDGLKSTFVPEGTNAVTADQSKSEASGSQAKI----------TSKPENL 585
KS D + +K TN + +S KI + L
Sbjct: 1219 NLK-KSEIDALNLQIKELKKKNETNEASLLESIKSVESETVKIKELQDECNFKEKEVSEL 1277
Query: 586 KIKVMTKSDPKSQKITILKRSETVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPK 645
+ K+ D S+ + + K SE + + L +++ Q ++ + A EK+ + + K
Sbjct: 1278 EDKLKASEDKNSKYLELQKESEKIKEELDAKTTELKIQLEKITNLSKAKEKSESELSRLK 1337
Query: 646 VLNDQKTLSIHPKVQGRKSTI 666
+ ++ + +++ K+ I
Sbjct: 1338 KTSSEERKNAEEQLEKLKNEI 1358
Score = 51.6 bits (122), Expect = 2e-05
Identities = 104/563 (18%), Positives = 226/563 (39%), Gaps = 67/563 (11%)
Query: 108 RTEYMKMSDKST-AKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDDESIEEMYSR 166
+ E +++ ++T K SL AN E +K E A L E K+ + EE+
Sbjct: 1123 KEEKIQLEKEATETKQQLNSLRANLESLEKEHEDLAAQLKKYEEQIANKERQYNEEISQL 1182
Query: 167 FQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVH 226
+ S Q + +D + ++++ S + + D L +++L + +
Sbjct: 1183 NDEITSTQQENESIKKKNDELEGEVKAMKSTSEEQSNLKKSEIDALNLQIKELKKKNETN 1242
Query: 227 EMSLNEHETSKKSKSIALP----------------------SKGKTSKSSKAYKASESEE 264
E SL E S +S+++ + S+ K SK + K SE +
Sbjct: 1243 EASLLESIKSVESETVKIKELQDECNFKEKEVSELEDKLKASEDKNSKYLELQKESEKIK 1302
Query: 265 ESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFI 324
E D + E + + K+ L++ ++K S+ K + E++K N ++ +
Sbjct: 1303 EELDAKTTE-----LKIQLEKITNLSKAKEKSESELSRLKKTSSEERK---NAEEQLEKL 1354
Query: 325 VDCPDLQKEKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEAD--DDAKAA 382
+ ++ + F+ + + SS ++ + + ++L ++ +A D+ ++
Sbjct: 1355 KNEIQIKNQAFEKERKLLNEGSSTITQEYSEKINTLEDELIRLQNENELKAKEIDNTRSE 1414
Query: 383 VGLVATVSSEAVSEAESDSED-ENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKY 441
+ V+ + E + E ++ + ++E+ S + ++ + ++LLS+ +L LKE+
Sbjct: 1415 LEKVSLSNDELLEEKQNTIKSLQDEILSY--KDKITRNDEKLLSIERDNKRDLESLKEQL 1472
Query: 442 --------------VDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKC 487
L ++ EL+ SKE +K + E LKS + +++
Sbjct: 1473 RAAQESKAKVEEGLKKLEEESSKEKAELEKSKEMMKKLESTIESNETELKSSMETIRKSD 1532
Query: 488 DKGSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSK---EYSLKSYCD 544
+K +K + A +D ++S + S I + K+ E KSK E S +
Sbjct: 1533 EKLEQSK-KSAEEDIKNLQHEKSDLISRINESEKD------IEELKSKLRIEAKSGSELE 1585
Query: 545 CIKDGLKSTFVPEGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTKSDPKSQKITILK 604
+K L + NA KS+ + ++ K +K + +K +
Sbjct: 1586 TVKQELNNAQEKIRINAEENTVLKSKLEDIERELKDKQAEIK-------SNQEEKELLTS 1638
Query: 605 RSETVHQNLIKPESKIPKQKDQK 627
R + + Q L + K K ++++
Sbjct: 1639 RLKELEQELDSTQQKAQKSEEER 1661
Score = 43.9 bits (102), Expect = 0.004
Identities = 91/430 (21%), Positives = 167/430 (38%), Gaps = 39/430 (9%)
Query: 89 KKLYKKHHKIRGIIVASIPRTE-----YMKMSDKSTAKAMFASLCANFEGSKKVKEAKAL 143
K+L KK+ ++ SI E ++ D+ K S + + + K +K L
Sbjct: 1233 KELKKKNETNEASLLESIKSVESETVKIKELQDECNFKEKEVSELEDKLKASEDKNSKYL 1292
Query: 144 MLVHQYELFRMKDDESIEEMYSRFQTLV----------SGLQILKK-SYVASDHVSKILR 192
L + E + + D E+ + + + S L LKK S + + L
Sbjct: 1293 ELQKESEKIKEELDAKTTELKIQLEKITNLSKAKEKSESELSRLKKTSSEERKNAEEQLE 1352
Query: 193 SLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSK 252
L + + K A E+ + L + +++ E E + L +K +
Sbjct: 1353 KLKNEIQIKNQAFEKERKLLNEGSSTITQEYS-EKINTLEDELIRLQNENELKAKEIDNT 1411
Query: 253 SSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKN---SKKE 309
S+ K S S +E + + +S++ +LS K + + R +K LS K S KE
Sbjct: 1412 RSELEKVSLSNDELLEEKQNTIKSLQDEILSYK-DKITRNDEKLLSIERDNKRDLESLKE 1470
Query: 310 DQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFRKQIK------KSLMATWED 363
+ K + + + K K + KS K I+ KS M T
Sbjct: 1471 QLRAAQESKAKVEEGLKKLEEESSKEKAELEKSKEMMKKLESTIESNETELKSSMETIRK 1530
Query: 364 LDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQEL----VDS 419
D + K+ A++D K + S+ +S +D E+ SK+ + +++
Sbjct: 1531 SDEKLEQSKKSAEEDIKN----LQHEKSDLISRINESEKDIEELKSKLRIEAKSGSELET 1586
Query: 420 LKELLSLFEHR----TNELTDLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDR 475
+K+ L+ + + E T LK K D+ ++ K E+K+++EE + E
Sbjct: 1587 VKQELNNAQEKIRINAEENTVLKSKLEDIERELKDKQAEIKSNQEEKELLTSRLKELEQE 1646
Query: 476 LKSLCQKLQE 485
L S QK Q+
Sbjct: 1647 LDSTQQKAQK 1656
Score = 40.0 bits (92), Expect = 0.054
Identities = 73/356 (20%), Positives = 138/356 (38%), Gaps = 25/356 (7%)
Query: 56 DEELWDILEDGVDDLDLDEEGAAIDRRIHTPAQ---KKLYKKHHKIRGIIVASIPRTEYM 112
DE+L I D DL+ +E + + KKL ++ K + + S + +
Sbjct: 1451 DEKLLSIERDNKRDLESLKEQLRAAQESKAKVEEGLKKLEEESSKEKAELEKSKEMMKKL 1510
Query: 113 KMSDKSTAKAMFASLCANFEGSKKVKEAKALMLVHQYELFRMKDD--ESIEEMYSRFQTL 170
+ + +S + +S+ + +K++++K L K D I E + L
Sbjct: 1511 ESTIESNETELKSSMETIRKSDEKLEQSKKSAEEDIKNLQHEKSDLISRINESEKDIEEL 1570
Query: 171 VSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSL 230
S L+I KS + V + L + + K+ E + +ED+ LK + +
Sbjct: 1571 KSKLRIEAKSGSELETVKQELNNA----QEKIRINAEENTVLKSKLEDIERELKDKQAEI 1626
Query: 231 NEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLA 290
++ K+ + L + S++ KA +SEEE V+ + L K L
Sbjct: 1627 KSNQEEKELLTSRLKELEQELDSTQQ-KAQKSEEERRA--EVRKFQVEKSQLDEKAMLLE 1683
Query: 291 RKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFR 350
K ++K ++K + +K + ++ + L KE K+ S +
Sbjct: 1684 TKYNDLVNKEQAWKRDEDTVKKTTDSQRQ------EIEKLAKELDNLKAENSKLKEANED 1737
Query: 351 KQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENE 406
+ LM DLD ++ + + D L +SS+ + E D EDE E
Sbjct: 1738 RSEIDDLMLLVTDLDEKNAKYRSKLKD-------LGVEISSDEEDDEEDDEEDEEE 1786
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog
(X-linked nuclear protein-1)
Length = 1359
Score = 57.0 bits (136), Expect = 4e-07
Identities = 75/286 (26%), Positives = 119/286 (41%), Gaps = 55/286 (19%)
Query: 231 NEHETSKKSKSIALPSKGKTSKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLA 290
+E E KKSKS + K KS K S SE+E DSDE++ K
Sbjct: 96 DEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSEDE----DSDEEREQKSK---------- 141
Query: 291 RKQKKFLSKRGSYKNSKKEDQKGCFNCKKPGHFIVDCPDLQKEKFKGKSMKSSFNSSKFR 350
+K KK + S + + E+++ KK KEK K ++S S +
Sbjct: 142 KKSKKTKKQTSSESSEESEEERKVKKSKK-----------NKEKSVKKRAETSEESDEDE 190
Query: 351 KQIKKSLMATWEDLDSESGSDKE--------------------EADDDA----KAAVGLV 386
K KKS + SES S+ E E++D+A K
Sbjct: 191 KPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRKR 250
Query: 387 ATVSSEAVSEAE-SDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLM 445
+ SSE SE+E SD E+E + S P+++ ++K+L S E +++ L +K
Sbjct: 251 SKTSSEESSESEKSDEEEEEKESSPKPKKKKPLAVKKLSSDEESEESDVEVLPQK----K 306
Query: 446 KQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGS 491
K+ TL+ +++ K + S E+++ K QE + GS
Sbjct: 307 KRGAVTLISDSEDEKDQKSESEASDV-EEKVSKKKAKKQESSESGS 351
Score = 54.3 bits (129), Expect = 3e-06
Identities = 92/435 (21%), Positives = 165/435 (37%), Gaps = 50/435 (11%)
Query: 207 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEES 266
E D + + EDL + ++ + K K + +K KAS SEE+
Sbjct: 9 EDSDGHVIEDEDLEMARQIENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSEEDD 68
Query: 267 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKKEDQKGCFNCKKP----GH 322
D + +S K + K E + + + ++ S K+ KK DQK KK
Sbjct: 69 DDEEESPRKSSKKSRKRAKSESESDESDEEEDRKKS-KSKKKVDQKKKEKSKKKRTTSSS 127
Query: 323 FIVDCPDLQKEKFKGKSMKS-------SFNSSKFRKQIKKSLMATWEDLDSESGSDKEEA 375
D + +++K K KS K+ S S+ +++KKS E + EE+
Sbjct: 128 EDEDSDEEREQKSKKKSKKTKKQTSSESSEESEEERKVKKS-KKNKEKSVKKRAETSEES 186
Query: 376 DDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELT 435
D+D K + + +A SE+ES+SEDE EV
Sbjct: 187 DEDEKPSKKSKKGLKKKAKSESESESEDEKEVKKS------------------------- 221
Query: 436 DLKEKYVDLMKQQKSTLLELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKH 495
K+K ++K++ + E K+ K +++ E + +E+ + K
Sbjct: 222 --KKKSKKVVKKESESEDEAPEKKKTEKRKRSKTSSEESSESEKSDEEEEEKESSPKPKK 279
Query: 496 EIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFV 555
+ L ++ + S+ + + K K + + E KS + ++ V
Sbjct: 280 KKPLAVKKLSSDEESEESDVEVLPQKKKRGAVTLISDSEDEKDQKSESE-------ASDV 332
Query: 556 PEGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTKSDPKSQKITI---LKRSETVHQN 612
E + A + +S SGS + S N K K K + K + I + + ET+
Sbjct: 333 EEKVSKKKAKKQESSESGSDSSEGSITVNRKSKKKEKPEKKKKGIIMDSSKLQKETIDAE 392
Query: 613 LIKPESKIPKQKDQK 627
+ E + +K QK
Sbjct: 393 RAEKERRKRLEKKQK 407
Score = 44.3 bits (103), Expect = 0.003
Identities = 86/417 (20%), Positives = 162/417 (38%), Gaps = 50/417 (11%)
Query: 262 SEEESPDGDSDEDQSVKMA--MLSNKLEYLARK------------QKKFLSKRGSYKNSK 307
SE E DG ED+ ++MA + + + E A+K KK +K+ +S+
Sbjct: 6 SESEDSDGHVIEDEDLEMARQIENERKEKRAQKLKEKREREGKPPPKKRPAKKRKASSSE 65
Query: 308 KED-------QKGCFNCKKPGHFIVDCPDLQKEKFKGKS-MKSSFNSSKFRKQIKKSLMA 359
++D +K +K + + +E+ + KS K + K K KK +
Sbjct: 66 EDDDDEEESPRKSSKKSRKRAKSESESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTS 125
Query: 360 TWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDS 419
+ ED DS+ +++ K + SE+ +SE+E +V K ++ S
Sbjct: 126 SSEDEDSDEEREQKSKKKSKK--------TKKQTSSESSEESEEERKV--KKSKKNKEKS 175
Query: 420 LKELLSLFEHRTNELTDLKEKYVDLMKQQKS-----TLLELKASKEELKGFNLISTTYED 474
+K+ E + K+ L K+ KS + E + K + K ++ E
Sbjct: 176 VKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESES 235
Query: 475 RLKSLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKS 534
++ +K EK + + E + + + + + S S K K + + S
Sbjct: 236 EDEAPEKKKTEKRKRSKTSSEESSESE--KSDEEEEEKES---SPKPKKKKPLAVKKLSS 290
Query: 535 KEYSLKSYCDCIKDGLKSTFVP--EGTNAVTADQSKSEASGSQAKITSKPENLKIKVMTK 592
E S +S + + K V + +S+SEAS + K++ K + +
Sbjct: 291 DEESEESDVEVLPQKKKRGAVTLISDSEDEKDQKSESEASDVEEKVSKKKAKKQESSESG 350
Query: 593 SDPKSQKITILKRS------ETVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVK 643
SD IT+ ++S E + +I SK+ K+ +A K + K K
Sbjct: 351 SDSSEGSITVNRKSKKKEKPEKKKKGIIMDSSKLQKETIDAERAEKERRKRLEKKQK 407
Score = 42.7 bits (99), Expect = 0.008
Identities = 71/340 (20%), Positives = 130/340 (37%), Gaps = 27/340 (7%)
Query: 333 EKFKGKSMKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSE 392
+K K K + K R K+ ++ ED D E S ++ + K
Sbjct: 37 QKLKEKREREGKPPPKKRPAKKRKASSSEEDDDDEEESPRKSSKKSRK-----------R 85
Query: 393 AVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTL 452
A SE+ESD DE E K ++ VD K+ S + T+ D + D ++QKS
Sbjct: 86 AKSESESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRTTSSSED---EDSDEEREQKSKK 142
Query: 453 LELKASKEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRSKV 512
K K+ + + E+R +K +EK K E + +D K
Sbjct: 143 KSKKTKKQTSSESS--EESEEERKVKKSKKNKEKSVKKRAETSEESDED----EKPSKKS 196
Query: 513 ASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAVTADQSKSEAS 572
+ K++ + E++ K+ KS K+ PE T + +S+ S
Sbjct: 197 KKGLKKKAKSESESESEDEKEVKKSKKKSKKVVKKESESEDEAPE--KKKTEKRKRSKTS 254
Query: 573 GSQAKITSKPENLKIKVMTKSDPKSQKITILKRSETVHQNLIKPESKIPKQKDQKNKAAT 632
++ + K + + + + PK +K +K+ + ++ +P QK ++
Sbjct: 255 SEESSESEKSDEEEEEKESSPKPKKKKPLAVKKLSSDEESEESDVEVLP-QKKKRGAVTL 313
Query: 633 ASEKTIPKGVKPKVLNDQKTLSIHPKVQGRKSTIGNSSIS 672
S+ K K ++ + + KV +K+ SS S
Sbjct: 314 ISDSEDEKDQK----SESEASDVEEKVSKKKAKKQESSES 349
Score = 40.0 bits (92), Expect = 0.054
Identities = 42/163 (25%), Positives = 73/163 (44%), Gaps = 20/163 (12%)
Query: 1000 AKAQRGIFLGYSERSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSED 1059
+K+++ + E+SK R +S E+S D E E K+ ++S+ T SE
Sbjct: 104 SKSKKKVDQKKKEKSKKKRTTSSSED--EDS------DEEREQKSKKKSKKTKKQTSSES 155
Query: 1060 ASESDQPSDSEKYTE-VESSPEAEITPEAESNSEAEPSPKVQN------ESASEDFQDNT 1112
+ ES++ +K + E S + ES+ + +PS K + +S SE ++
Sbjct: 156 SEESEEERKVKKSKKNKEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESESEDE 215
Query: 1113 QQVIQPKFK-----HKSSHHEELIIGSKDSPRRTRSHFRQEES 1150
++V + K K K S E+ K + +R RS EES
Sbjct: 216 KEVKKSKKKSKKVVKKESESEDEAPEKKKTEKRKRSKTSSEES 258
Score = 37.7 bits (86), Expect = 0.27
Identities = 31/170 (18%), Positives = 73/170 (42%), Gaps = 19/170 (11%)
Query: 1013 RSKAYRVYNSETQCVEESMHVKFDDREPESKTSEQSESNAGTTDSEDASESDQPSDSEK- 1071
+ K+ +V E++ +E+ K ++ SKTS + S + +D E+ + P +K
Sbjct: 222 KKKSKKVVKKESESEDEAPEKKKTEKRKRSKTSSEESSESEKSDEEEEEKESSPKPKKKK 281
Query: 1072 --------YTEVESSPEAEITPEAESN------SEAEPSPKVQNESASEDFQDNTQQVIQ 1117
E + E+ P+ + S++E ++ES + D ++ + +
Sbjct: 282 PLAVKKLSSDEESEESDVEVLPQKKKRGAVTLISDSEDEKDQKSESEASDVEEKVSKK-K 340
Query: 1118 PKFKHKSSHHEELIIGSKDSPRRTRSHFRQEESLIGLL---SIIEPKTVE 1164
K + S + GS R+++ + E+ G++ S ++ +T++
Sbjct: 341 AKKQESSESGSDSSEGSITVNRKSKKKEKPEKKKKGIIMDSSKLQKETID 390
>IF38_SCHPO (O14164) Probable eukaryotic translation initiation factor
3 93 kDa subunit (eIF3 p93)
Length = 918
Score = 55.5 bits (132), Expect = 1e-06
Identities = 57/204 (27%), Positives = 87/204 (41%), Gaps = 35/204 (17%)
Query: 1007 FLGYSERSKAYRVYNSETQCVEESMHVKFDDR-------EPESKTSEQSESNAGTTDSED 1059
F G S S A V +SE + S K DD E ES +S +SES+ ++SE+
Sbjct: 5 FKGGSSDSDAESVDSSEENRLTSSRLKKQDDSSSEEESSEEESASSSESESSEEESESEE 64
Query: 1060 A---------------SESDQPSDSEKYTEVESSPEAEITPEAESNSEAEPSPKVQNESA 1104
+ SESD S SE+ E ES ++E++ E+ES SE+E + ES
Sbjct: 65 SEVEVPKKKAVAASEDSESDSES-SEEEEETESEEDSEVSDESESESESESES--EEESE 121
Query: 1105 SEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSHFRQEESLIGLLSIIEPK--- 1161
SE+ D +++ F K E G K + EE ++ + K
Sbjct: 122 SEEESDESERSGPSSFLKKPEKEEAKPAGLKFLRGESSEESSDEEEGRRVVKSAKDKRYE 181
Query: 1162 -------TVEEALSDDGWILAMQE 1178
T++ A+S + WI+ E
Sbjct: 182 EFISCMETIKNAMSSNNWIVVSNE 205
Score = 36.2 bits (82), Expect = 0.78
Identities = 36/148 (24%), Positives = 66/148 (44%), Gaps = 16/148 (10%)
Query: 354 KKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSK--- 410
KK +A ED +S+S S +EE + +++ + SE+ SE+ES+ E E+E S
Sbjct: 71 KKKAVAASEDSESDSESSEEEEETESEEDSEVSDESESESESESESEEESESEEESDESE 130
Query: 411 -------IPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLE-----LKAS 458
+ + E ++ L ++E + +E+ ++K K E ++
Sbjct: 131 RSGPSSFLKKPEKEEAKPAGLKFLRGESSEESSDEEEGRRVVKSAKDKRYEEFISCMETI 190
Query: 459 KEELKGFNLISTTYE-DRLKSLCQKLQE 485
K + N I + E D L + QK +E
Sbjct: 191 KNAMSSNNWIVVSNEFDHLNKVSQKCKE 218
Score = 34.7 bits (78), Expect = 2.3
Identities = 29/108 (26%), Positives = 47/108 (42%), Gaps = 1/108 (0%)
Query: 206 EEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKTSKSSKAYKASESEEE 265
EE+ + + S + SS + E +E E KK A S+SS+ + +ESEE+
Sbjct: 40 EESSEEESASSSESESSEEESESEESEVEVPKKKAVAASEDSESDSESSEEEEETESEED 99
Query: 266 SPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSY-KNSKKEDQK 312
S D E +S + + E + S S+ K +KE+ K
Sbjct: 100 SEVSDESESESESESESEEESESEEESDESERSGPSSFLKKPEKEEAK 147
Score = 34.3 bits (77), Expect = 3.0
Identities = 53/219 (24%), Positives = 93/219 (42%), Gaps = 39/219 (17%)
Query: 1030 SMHVKFDDREPESKTSEQSESNAGTTD---SEDASESDQPSDSEKYTEVESSPEAEITPE 1086
S K + ++++ + SE N T+ +D S S++ S E E SS E+E + E
Sbjct: 2 SRFFKGGSSDSDAESVDSSEENRLTSSRLKKQDDSSSEEESSEE---ESASSSESESSEE 58
Query: 1087 AESNSEAEPS-PKVQNESASEDFQDNTQQVIQPKFKHKSSHHEELIIGSKDSPRRTRSHF 1145
+ E+E PK + +ASED + +++ SS EE +DS S
Sbjct: 59 ESESEESEVEVPKKKAVAASEDSESDSE----------SSEEEEETESEEDSEVSDESES 108
Query: 1146 RQEESLIGLLSIIEPKTVEEALSDDGWILAMQEELNQFQRNDVWDLVPKPFQKNI--IGT 1203
E E ++ EE+ S +EE ++ +R+ + KP ++ G
Sbjct: 109 ESES---------ESESEEESES--------EEESDESERSGPSSFLKKPEKEEAKPAGL 151
Query: 1204 KWVFRNKLNEQGEVTRNKARLV--AQGYSQQEGIDYTET 1240
K++ R + +E+ R+V A+ +E I ET
Sbjct: 152 KFL-RGESSEESSDEEEGRRVVKSAKDKRYEEFISCMET 189
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.131 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 192,102,984
Number of Sequences: 164201
Number of extensions: 8342931
Number of successful extensions: 30318
Number of sequences better than 10.0: 663
Number of HSP's better than 10.0 without gapping: 169
Number of HSP's successfully gapped in prelim test: 519
Number of HSP's that attempted gapping in prelim test: 26439
Number of HSP's gapped (non-prelim): 3079
length of query: 1667
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1543
effective length of database: 39,613,130
effective search space: 61123059590
effective search space used: 61123059590
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 73 (32.7 bits)
Medicago: description of AC147435.7


