

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147435.10 + phase: 0 /pseudo
(612 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
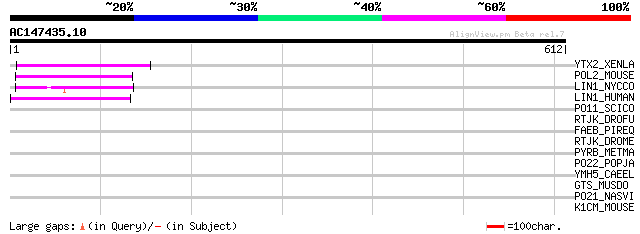
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 77 2e-13
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 62 4e-09
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 58 8e-08
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 52 6e-06
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 40 0.014
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 36 0.34
FAEB_PIREQ (Q9Y871) Feruloyl esterase B precursor (EC 3.1.1.73) ... 35 0.75
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 33 1.7
PYRB_METMA (Q8PXK5) Aspartate carbamoyltransferase (EC 2.1.3.2) ... 33 1.7
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 33 1.7
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 33 2.9
GTS_MUSDO (P46437) Glutathione S-transferase (EC 2.5.1.18) (GST ... 32 4.9
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 32 6.4
K1CM_MOUSE (P08730) Keratin, type I cytoskeletal 13 (Cytokeratin... 32 6.4
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 76.6 bits (187), Expect = 2e-13
Identities = 43/148 (29%), Positives = 77/148 (51%)
Query: 8 LSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIH 67
+SE + L P ++E+ +R NKSPG D F + FW + + + +
Sbjct: 437 VSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAF 496
Query: 68 ANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVIST 127
LP S ++L+PK +K++R +SLL + YK++AK ++ RL VL+ VI
Sbjct: 497 KKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHP 556
Query: 128 TQSAFLKGTNLVDGVLVVNELVDYAKKS 155
QS + G + D V ++ +L+ +A+++
Sbjct: 557 DQSYTVPGRTIFDNVFLIRDLLHFARRT 584
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 62.0 bits (149), Expect = 4e-09
Identities = 39/130 (30%), Positives = 66/130 (50%), Gaps = 1/130 (0%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQI 66
+L++ D L +P +EIE V+ KSPG D ++ F + F + + +F +I
Sbjct: 466 KLNQDQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKI 525
Query: 67 HANELLPKSMLAYFVALIPKVSS-PLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVI 125
LP S + LIPK P +++++R ISL+ K+L K+LA R+ + ++I
Sbjct: 526 EVEGTLPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAII 585
Query: 126 STTQSAFLKG 135
Q F+ G
Sbjct: 586 HPDQVGFIPG 595
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 57.8 bits (138), Expect = 8e-08
Identities = 40/135 (29%), Positives = 62/135 (45%), Gaps = 9/135 (6%)
Query: 7 RLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEV----RIM 62
RLS+ + ++L P EI ++ KSPG D F EF+ K E+ +
Sbjct: 439 RLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPD----GFTSEFYQTFKEELVPILLNL 494
Query: 63 FDQIHANELLPKSMLAYFVALIPKVSS-PLELKDYRTISLLGSLYKLLAKVLARRLAGVL 121
F I +LP + + LIPK P ++YR ISL+ K+L K+L R+ +
Sbjct: 495 FQNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHI 554
Query: 122 SSVISTTQSAFLKGT 136
+I Q F+ G+
Sbjct: 555 KKIIHHDQVGFIPGS 569
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 51.6 bits (122), Expect = 6e-06
Identities = 35/134 (26%), Positives = 60/134 (44%), Gaps = 1/134 (0%)
Query: 1 DGVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVR 60
D RL++ + + L P EIE ++ KSPG + + F + + + +
Sbjct: 433 DTYTLPRLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLL 492
Query: 61 IMFDQIHANELLPKSMLAYFVALIPKVSSPLELKD-YRTISLLGSLYKLLAKVLARRLAG 119
+F I +LP S + LIPK K+ +R ISL+ K+L K+LA ++
Sbjct: 493 KLFQSIEKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQ 552
Query: 120 VLSSVISTTQSAFL 133
+ +I Q F+
Sbjct: 553 HIKKLIHHDQVGFI 566
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 40.4 bits (93), Expect = 0.014
Identities = 40/152 (26%), Positives = 64/152 (41%), Gaps = 12/152 (7%)
Query: 9 SEVDNDLLVA-----PFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMF 63
SE+D +A M+E+ VR KSPG D V+ W + + ++
Sbjct: 389 SEIDRLKAIARPLPPDLEMDEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLY 448
Query: 64 DQIHANELLPK----SMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAG 119
Q P+ + L + L+ ++ S + YR I LL +L K+L ++ +RL
Sbjct: 449 KQCLLESYFPQKWKIASLVILLKLLDRIRS--DPGSYRPICLLDNLGKVLEGIMVKRLDQ 506
Query: 120 VLSSV-ISTTQSAFLKGTNLVDGVLVVNELVD 150
L V +S Q AF G + D V V+
Sbjct: 507 KLMDVEVSPYQFAFTYGKSTEDAWRCVQRHVE 538
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 35.8 bits (81), Expect = 0.34
Identities = 25/100 (25%), Positives = 49/100 (49%), Gaps = 1/100 (1%)
Query: 19 PFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKSM-L 77
P +EE++ +V + K+PG D + ++ + ++F+ I PK+
Sbjct: 437 PVTLEEVKELVSKLKPKKAPGEDLLDNRTIRLLPDQALLYLVLIFNSILRVGYFPKARPT 496
Query: 78 AYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRL 117
A + ++ PL++ YR SLL SL K+L +++ R+
Sbjct: 497 ASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRI 536
>FAEB_PIREQ (Q9Y871) Feruloyl esterase B precursor (EC 3.1.1.73)
(Ferulic acid esterase B) (Esterase A) (EstA) (Cinnamoyl
ester hydrolase)
Length = 536
Score = 34.7 bits (78), Expect = 0.75
Identities = 22/51 (43%), Positives = 26/51 (50%), Gaps = 13/51 (25%)
Query: 551 GGYGGGL-WGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGGNKMVR*GG 600
G GGG+ WGDFGG G GGG+ P+G FGGN+ GG
Sbjct: 116 GNQGGGMPWGDFGGNQGGN---------QGGGM---PWGDFGGNQGGNQGG 154
Score = 33.9 bits (76), Expect = 1.3
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 13/45 (28%)
Query: 551 GGYGGGL-WGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGGNK 594
G GGG+ WGDFGG G GGG+ P+G FGGN+
Sbjct: 133 GNQGGGMPWGDFGGNQGGN---------QGGGM---PWGDFGGNQ 165
Score = 33.9 bits (76), Expect = 1.3
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 13/45 (28%)
Query: 551 GGYGGGL-WGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGGNK 594
G GGG+ WGDFGG G GGG+ P+G FGGN+
Sbjct: 193 GNQGGGMQWGDFGGNQGGN---------QGGGM---PWGDFGGNQ 225
Score = 33.9 bits (76), Expect = 1.3
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 13/45 (28%)
Query: 551 GGYGGGL-WGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGGNK 594
G GGG+ WGDFGG G GGG+ P+G FGGN+
Sbjct: 163 GNQGGGMQWGDFGGNQGGN---------QGGGM---PWGDFGGNQ 195
Score = 33.5 bits (75), Expect = 1.7
Identities = 22/51 (43%), Positives = 26/51 (50%), Gaps = 4/51 (7%)
Query: 551 GGYGGGL-WGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGGNKMVR*GG 600
G GGG+ WGDFGG G GGG+ P+G FGGN+ GG
Sbjct: 90 GNQGGGMQWGDFGGNQGGGMPWGDFGGNQGGGM---PWGDFGGNQGGNQGG 137
Score = 33.5 bits (75), Expect = 1.7
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 4/45 (8%)
Query: 551 GGYGGGL-WGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGGNK 594
G GGG+ WGDFGG G GGG+ P+G FGGN+
Sbjct: 77 GNQGGGMPWGDFGGNQGGGMQWGDFGGNQGGGM---PWGDFGGNQ 118
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 33.5 bits (75), Expect = 1.7
Identities = 22/97 (22%), Positives = 49/97 (49%), Gaps = 1/97 (1%)
Query: 22 MEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKSMLAYFV 81
+EE++ ++ + K+PG D + ++ + ++F+ + PK+ + +
Sbjct: 440 LEEVKNLIAKLPLKKAPGEDLLDNRTIRLLPDQALQFLALIFNSVLDVGYFPKAWKSASI 499
Query: 82 ALIPKVS-SPLELKDYRTISLLGSLYKLLAKVLARRL 117
+I K +P ++ YR SLL SL K++ +++ RL
Sbjct: 500 IMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRL 536
>PYRB_METMA (Q8PXK5) Aspartate carbamoyltransferase (EC 2.1.3.2)
(Aspartate transcarbamylase) (ATCase)
Length = 309
Score = 33.5 bits (75), Expect = 1.7
Identities = 22/62 (35%), Positives = 34/62 (54%), Gaps = 4/62 (6%)
Query: 93 LKDYRTISLL----GSLYKLLAKVLARRLAGVLSSVISTTQSAFLKGTNLVDGVLVVNEL 148
L D + ISLL + +L +V RRL G + S+ S S+ +KG NL D + V+++
Sbjct: 42 LLDGKIISLLFFEPSTRTRLSFEVATRRLGGQVLSLGSVEASSVMKGENLADTIRVISKY 101
Query: 149 VD 150
D
Sbjct: 102 AD 103
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 33.5 bits (75), Expect = 1.7
Identities = 35/132 (26%), Positives = 61/132 (45%), Gaps = 12/132 (9%)
Query: 19 PFHMEEIEVVVRESDGNKSPGADEYNFAFVKEFWYLMKNEVRIMFDQIHANELLPKSMLA 78
P EEI+ ++ + +PG+D + L +N V++ + H +P A
Sbjct: 5 PIAREEIQCAIKGWKPS-APGSDGLTVQAITRT-RLPRNFVQLHLLRGH----VPTPWTA 58
Query: 79 YFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSVISTTQSAFLKGTNL 138
LIPK ++R I++ +L +LL ++LA+RL + + Q KG
Sbjct: 59 MRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQ----KGYAR 112
Query: 139 VDGVLVVNELVD 150
+DG LV + L+D
Sbjct: 113 IDGTLVNSLLLD 124
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 32.7 bits (73), Expect = 2.9
Identities = 17/69 (24%), Positives = 32/69 (45%)
Query: 65 QIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAKVLARRLAGVLSSV 124
Q ++ +P + IPK +P +YR ISL +++ +++ R+ S +
Sbjct: 654 QSFSDSAIPNRWKHAVIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHL 713
Query: 125 ISTTQSAFL 133
+S Q FL
Sbjct: 714 LSPHQHGFL 722
>GTS_MUSDO (P46437) Glutathione S-transferase (EC 2.5.1.18) (GST
class-sigma)
Length = 241
Score = 32.0 bits (71), Expect = 4.9
Identities = 17/63 (26%), Positives = 36/63 (56%), Gaps = 6/63 (9%)
Query: 1 DGVNFNRLSEVDNDLLVAPFHMEEIEVVVRESDG----NKSPGADEYNFAFVKEFWYLMK 56
D + +L ++N+++ PF++E++E V+++DG NK AD Y + Y++K
Sbjct: 149 DEIKEKKLVTLNNEVI--PFYLEKLEQTVKDNDGHLALNKLTWADVYFAGILDYMNYMVK 206
Query: 57 NEV 59
++
Sbjct: 207 RDI 209
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 31.6 bits (70), Expect = 6.4
Identities = 19/102 (18%), Positives = 46/102 (44%), Gaps = 2/102 (1%)
Query: 52 WYLMKNEVRIMFDQIHANELLPKSMLAYFVALIPKVSSPLELKDYRTISLLGSLYKLLAK 111
W + ++ +F+ I + P+ L LIPK ++ +R +S+ + +
Sbjct: 345 WNSIDECIKSLFNMIMYHGQCPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHR 404
Query: 112 VLARRLAGVLSSVISTTQSAFLKGTNLVDGVLVVNELVDYAK 153
+LA R+ ++ T Q AF+ + + +++ ++ A+
Sbjct: 405 ILANRIGE--HGLLDTRQRAFIVADGVAENTSLLSAMIKEAR 444
>K1CM_MOUSE (P08730) Keratin, type I cytoskeletal 13 (Cytokeratin
13) (K13) (CK 13) (47 kDa cytokeratin)
Length = 437
Score = 31.6 bits (70), Expect = 6.4
Identities = 19/44 (43%), Positives = 21/44 (47%), Gaps = 14/44 (31%)
Query: 549 SNGGYGGGLWGDFGGELGLEFYLEAKTICMGGGVAY*PFGRFGG 592
+ GG+GGG G FGG G F GGG FG FGG
Sbjct: 57 AGGGFGGGFGGGFGGSYGGGF---------GGG-----FGDFGG 86
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.358 0.165 0.605
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,056,385
Number of Sequences: 164201
Number of extensions: 2300656
Number of successful extensions: 10963
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 10922
Number of HSP's gapped (non-prelim): 30
length of query: 612
length of database: 59,974,054
effective HSP length: 116
effective length of query: 496
effective length of database: 40,926,738
effective search space: 20299662048
effective search space used: 20299662048
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147435.10


