

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147434.12 - phase: 0
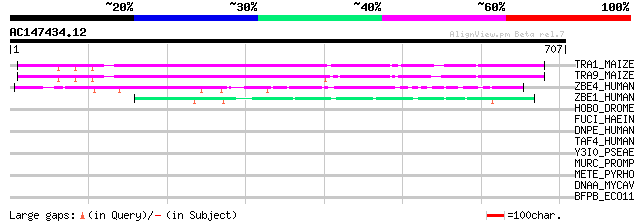
(707 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
TRA1_MAIZE (P08770) Putative AC transposase (ORFA) 281 3e-75
TRA9_MAIZE (P03010) Putative AC9 transposase 266 2e-70
ZBE4_HUMAN (O75132) Zinc finger BED domain containing protein 4 108 4e-23
ZBE1_HUMAN (O96006) Zinc finger BED domain containing protein 1 ... 59 3e-08
HOBO_DROME (P12258) Transposable element Hobo transposase 39 0.047
FUCI_HAEIN (P44779) L-fucose isomerase (EC 5.3.1.25) 36 0.40
DNPE_HUMAN (Q9ULA0) Aspartyl aminopeptidase (EC 3.4.11.21) 33 2.0
TAF4_HUMAN (O00268) Transcription initiation factor TFIID subuni... 33 3.4
Y3I0_PSEAE (Q51470) Hypothetical UPF0004 protein PA3980 32 4.4
MURC_PROMP (Q7V3P8) UDP-N-acetylmuramate--L-alanine ligase (EC 6... 32 5.8
METE_PYRHO (O58816) Probable methylcobalamin:homocysteine methyl... 32 7.5
DNAA_MYCAV (P49990) Chromosomal replication initiator protein dnaA 32 7.5
BFPB_ECO11 (Q9S142) Outer membrane lipoprotein bfpB precursor (B... 31 9.8
>TRA1_MAIZE (P08770) Putative AC transposase (ORFA)
Length = 806
Score = 281 bits (720), Expect = 3e-75
Identities = 205/700 (29%), Positives = 333/700 (47%), Gaps = 60/700 (8%)
Query: 10 QNLSSTLTSGSANVAEPTEQVAVATQVPVVGLPPLPCLAKRRKPNAGGPRR-----TSPA 64
Q++SS+ +G+A + V P P P + +P P++ TS
Sbjct: 83 QSVSSSNANGTATDPSQDDMAIVHEPQPQPQPQPEPQPQPQPEPEEEAPQKRAKKCTSDV 142
Query: 65 WDHFIKLPDEPEPTAACI----------HCHKRYLCDPKTHGTSNLLAH---------SK 105
W HF K E E +C +Y + HGTS H +
Sbjct: 143 WQHFTKKEIEVEVDGKKYVQVWGHCNFPNCKAKYRAEGH-HGTSGFRNHLRTSHSLVKGQ 201
Query: 106 VCFKNPQNDPTQASLMFSNGEGGTLVAASQRFNPAACRKAIALFVLLDEHAFRVVEGEGF 165
+C K+ ++ +L+ +++ K + L +++ E+ F +VE E F
Sbjct: 202 LCLKSEKDHGKDINLI-----------EPYKYDEVVSLKKLHLAIIMHEYPFNIVEHEYF 250
Query: 166 KLLCRQLQPLLTIPSRRTVARDCFQLFLDENLRLKTYFKSDCVRVALTTDCWTSGQNFSY 225
+ L+P I SR T + L+L+E +L K R + T D WTS QN SY
Sbjct: 251 VEFVKSLRPHFPIKSRVTARKYIMDLYLEEKEKLYGKLKDVQSRFSTTMDMWTSCQNKSY 310
Query: 226 MTLTAHFINNDWKYEKRILSFCTVPN-HKGDTIGRKVEEILKEWGI-RNVSTITVDNASS 283
M +T H+I++DW +KRI+ F V H G + + I+ +W I + + +++DNAS+
Sbjct: 311 MCVTIHWIDDDWCLQKRIVGFFHVEGRHTGQRLSQTFTAIMVKWNIEKKLFALSLDNASA 370
Query: 284 NDVAVAYLKKRINNM-GGLMGDGSFFHLRCCAHILNLVVGDGLKQNELSISSIRNSVRFV 342
N+VAV + + + + L+ DG+FFH+RC HILNLV DGL +I I+ V V
Sbjct: 371 NEVAVHDIIEDLQDTDSNLVCDGAFFHVRCACHILNLVAKDGLAVIAGTIEKIKAIVLAV 430
Query: 343 RSSPQRSAKFKECIEFARITCKKMLCLDVQTRWNTAYLMLDGAEKFQPAFEKLEGEDSGY 402
+SSP + + +C + K + DV TRWN+ YLML A ++PA +L+ D
Sbjct: 431 KSSPLQWEELMKCASECDLDKSKGISYDVSTRWNSTYLMLRDALYYKPALIRLKTSDPRR 490
Query: 403 LEFFGEAGPPSIHDWENVRCFVRFLKIFYDATKEFSSSQGVSLHKAFHQLASVHCELKRS 462
+ A P +W+ + LK F+D T+ S +Q + + + + + +
Sbjct: 491 YD----AICPKAEEWKMALTLFKCLKKFFDLTELLSGTQYSTANLFYKGFCEIKDLIDQW 546
Query: 463 AMNLNTVLASMGSDMKQKYNKYWGKIENINKLIYFGVILDPRYKFSYVEWCFNDMYGDQP 522
++ V+ M M +K+ KYW K+ NI + LDPRYK +E+ +GD
Sbjct: 547 CVHEKFVIRRMAVAMSEKFEKYW-KVSNIALAV--ACFLDPRYKKILIEFYMKKFHGDSY 603
Query: 523 TFFTDLIAMIHTQLFKLFNWYKDAYDQQHNSGHPSASPSESSYVSENVIPAEVPSHLARA 582
D + +L++ ++ S PSA P + ++++ + +
Sbjct: 604 KVHVDDFVRVIRKLYQFYS-----------SCSPSA-PKTKTTTNDSMDDTLMENEDDEF 651
Query: 583 EAFKEHLKLKESIVKKNELERYLDEERAEDVN-FDILLSWKQNSCRYPVLSSMVRDVLAT 641
+ + LK + V+ NEL++Y+ E + FDIL W+ YP+L+ + RDVLA
Sbjct: 652 QNYLHELKDYDQ-VESNELDKYMSEPLLKHSGQFDILSWWRGRVAEYPILTQIARDVLAI 710
Query: 642 PVSTVASESAFSTGGRVLDTYRSSLNPQMAEALICAQNWL 681
VSTVASESAFS GGRV+D YR+ L ++ EALIC ++W+
Sbjct: 711 QVSTVASESAFSAGGRVVDPYRNRLGSEIVEALICTKDWV 750
>TRA9_MAIZE (P03010) Putative AC9 transposase
Length = 839
Score = 266 bits (679), Expect = 2e-70
Identities = 207/725 (28%), Positives = 333/725 (45%), Gaps = 86/725 (11%)
Query: 10 QNLSSTLTSGSANVAEPTEQVAVATQVPVVGLPPLPCLAKRRKPNAGGPRR-----TSPA 64
Q++SS+ +G+A + V P P P + +P P++ TS
Sbjct: 29 QSVSSSNANGTATDPSQDDMAIVHEPQPQPQPQPEPQPQPQPEPEEEAPQKRAKKCTSDV 88
Query: 65 WDHFIKLPDEPEPTAACI----------HCHKRYLCDPKTHGTSNLLAH---------SK 105
W HF K E E +C +Y + HGTS H +
Sbjct: 89 WQHFTKKEIEVEVDGKKYVQVWGHCNFPNCKAKYRAEGH-HGTSGFRNHLRTSHSLVKGQ 147
Query: 106 VCFKNPQNDPTQASLMFSNGEGGTLVAASQRFNPAACRKAIALFVLLDEHAFRVVEGEGF 165
+C K+ ++ +L+ +++ K + L +++ E+ F +VE E F
Sbjct: 148 LCLKSEKDHGKDINLI-----------EPYKYDEVVSLKKLHLAIIMHEYPFNIVEHEYF 196
Query: 166 KLLCRQLQPLLTIPSRRTVARDCFQLFLDENLRLKTYFKSDCVRVALTTDCWTSGQNFSY 225
+ L+P I SR T + L+L+E +L K R + T D WTS QN SY
Sbjct: 197 VEFVKSLRPHFPIKSRVTARKYIMDLYLEEKEKLYGKLKDVQSRFSTTMDMWTSCQNKSY 256
Query: 226 MTLTAHFINNDWKYEKRILSFCTVPN-HKGDTIGRKVEEILKEWGI-RNVSTITVDNASS 283
M +T H+I++DW +KRI+ F V H G + + I+ +W I + + +++DNAS+
Sbjct: 257 MCVTIHWIDDDWCLQKRIVGFFHVEGRHTGQRLSQTFTAIMVKWNIEKKLFALSLDNASA 316
Query: 284 NDVAVAYLKKRINNM-GGLMGDGSFFHLRCCAHILNLVVGDGLKQNELSISSIRNSVRFV 342
N+VAV + + + + L+ DG+FFH+RC HILNLV DGL +I I+ V V
Sbjct: 317 NEVAVHDIIEDLQDTDSNLVCDGAFFHVRCACHILNLVAKDGLAVIAGTIEKIKAIVLAV 376
Query: 343 RSSPQRSAKFKECIEFARITCKKMLCLDVQTRWNTAYLMLDGAEKFQPAFEKLEGEDS-- 400
+SSP + + +C + K + DV TRWN+ YLML A ++PA +L+ D
Sbjct: 377 KSSPLQWEELMKCASECDLDKSKGISYDVSTRWNSTYLMLRDALYYKPALIRLKTSDPRR 436
Query: 401 -----------------------GYLEFFGEAGPPSIHDWENVRCFVRFLKIFYDATKEF 437
G ++F + P W + LK F+D T+
Sbjct: 437 YVCLNCCTCHHYKFSINQMSIIVGTMQFVLK---PRSGRWH--LTLFKCLKKFFDLTELL 491
Query: 438 SSSQGVSLHKAFHQLASVHCELKRSAMNLNTVLASMGSDMKQKYNKYWGKIENINKLIYF 497
S +Q + + + + + + + V+ M M +K+ KYW K+ NI +
Sbjct: 492 SGTQYSTANLFYKGFCEIKDLIDQWCCHEKFVIRRMAVAMSEKFEKYW-KVSNI--ALAV 548
Query: 498 GVILDPRYKFSYVEWCFNDMYGDQPTFFTDLIAMIHTQLFKLFNWYKDAYDQQHNSGHPS 557
LDPRYK +E+ +GD D + +L+ Q ++S PS
Sbjct: 549 ACFLDPRYKKILIEFYMKKFHGDSYKVHVDDFVRVIRKLY-----------QFYSSCSPS 597
Query: 558 ASPSESSYVSENVIPAEVPSHLARAEAFKEHLKLKESIVKKNELERYLDEERAE-DVNFD 616
A P + ++++ + + + + LK + V+ NEL++Y+ E + FD
Sbjct: 598 A-PKTKTTTNDSMDDTLMENEDDEFQNYLHELKDYDQ-VESNELDKYMSEPLLKHSGQFD 655
Query: 617 ILLSWKQNSCRYPVLSSMVRDVLATPVSTVASESAFSTGGRVLDTYRSSLNPQMAEALIC 676
IL W+ YP+L+ + RDVLA VSTVASESAFS GGRV+D YR+ L ++ EALIC
Sbjct: 656 ILSWWRGRVAEYPILTQIARDVLAIQVSTVASESAFSAGGRVVDPYRNRLGSEIVEALIC 715
Query: 677 AQNWL 681
++W+
Sbjct: 716 TKDWV 720
>ZBE4_HUMAN (O75132) Zinc finger BED domain containing protein 4
Length = 1171
Score = 108 bits (271), Expect = 4e-23
Identities = 161/688 (23%), Positives = 281/688 (40%), Gaps = 109/688 (15%)
Query: 7 SISQNLSSTLTSGSANVAEPTEQVAVATQVPVVGLPPLPCLAKRRKPNAGGPRRTSPAWD 66
S++ + +TL S ++ ++ T+ V T+ V P ++TS W+
Sbjct: 519 SLANSPYATLASAESSSSKLTDLPTVVTKNNQVMFPV-------------NSKKTSKLWN 565
Query: 67 HFIKLPDEPEPTAACIHCHKRYLCD--PKTHGTSNLLAHSKV----CFKNPQNDPTQASL 120
HF + C+HC + P GTS LL H + K ++ + S
Sbjct: 566 HF-SICSADSTKVVCLHCGRTISRGKKPTNLGTSCLLRHLQRFHSNVLKTEVSETARPSS 624
Query: 121 MFSNGEGGTLVAASQRFN----------PAACR--KAIALFVLLDEHAFRVVEGEGFKLL 168
+ GT ++ + F+ P A + IA + LD + V+ GF L
Sbjct: 625 PDTRVPRGTELSGASSFDDTNEKFYDSHPVAKKITSLIAEMIALDLQPYSFVDNVGFNRL 684
Query: 169 CRQLQPLLTIPSRRTVARDCFQLFLD--ENLRLKTYFKSDCVRVALTTDCWTSGQNFSYM 226
L+P ++P+ +R D + + + +++ + T+ W S Q Y+
Sbjct: 685 LEYLKPQYSLPAPSYFSRTAIPGMYDNVKQIIMSHLKEAESGVIHFTSGIWMSNQTREYL 744
Query: 227 TLTAHFINNDWKYEKR--------ILSFCTVP-NHKGDTIGRKVEEILKEW----GIRNV 273
TLTAH+++ + R +L V ++ G++I +++E + W G++
Sbjct: 745 TLTAHWVSFESPARPRCDDHHCSALLDVSQVDCDYSGNSIQKQLECWWEAWVTSTGLQVG 804
Query: 274 STITVDNASSNDVAVAYLKKRINNMGGLMGDGSFFHLRCCAHILNLVVGDGLK-----QN 328
T+T DNAS +G + +G ++C +H +NL+V + +K QN
Sbjct: 805 ITVT-DNAS---------------IGKTLNEGEHSSVQCFSHTVNLIVSEAIKSQRMVQN 848
Query: 329 ELSISSIRNSVRFVRSSPQRSAKFKECIEFARITCKKMLCLDVQTRWNTAYLMLDGAEKF 388
LS++ R V SP+ K E ++ + L DV ++W+T++ ML+ +
Sbjct: 849 LLSLA--RKICERVHRSPKAKEKLAE-LQREYALPQHHLIQDVPSKWSTSFHMLERLIEQ 905
Query: 389 QPAFEKLEGEDSGYLEFFGEAGPPSIHDWENVRCFVRFLKIFYDATKEFSSSQGV--SLH 446
+ A ++ E + E S WE ++ R LK F A++E S+ +
Sbjct: 906 KRAINEMSVE-CNFRELI------SCDQWEVMQSVCRALKPFEAASREMSTQMSTLSQVI 958
Query: 447 KAFHQLASVHCELKRSAMNLNTVLASMGSDMKQKYNKYWGKIENINKLIYFGVILDPRYK 506
H L L M ++T+L S+ M + + + F +LDPRYK
Sbjct: 959 PMVHILNRKVEMLFEETMGIDTMLRSLKEAMVSRLSATLHDPRYV-----FATLLDPRYK 1013
Query: 507 FSYVEWCFNDMYGDQPTFFTDLIAMIHTQLFKLFNWYKDAYDQQHNSGHPSASPSESSYV 566
S F + +Q + DLI + +L N + H + SPS+ S
Sbjct: 1014 AS----LFTEEEAEQ--YKQDLIREL-----ELMNSTSEDVAASHRC--DAGSPSKDSAA 1060
Query: 567 SENVIPAEVPSHLARAEAFKEHLKLKESIVKKNELERYLDEERAEDVNFDILLSWKQNSC 626
EN+ S +A+ + KL E++V YL+EE E + D L W
Sbjct: 1061 EENLW-----SLVAKVKKKDPREKLPEAMVL-----AYLEEEVLEH-SCDPLTYWNLKKA 1109
Query: 627 RYPVLSSMVRDVLATPVSTVASESAFST 654
+P LS++ L P S V SE F+T
Sbjct: 1110 SWPGLSALAVRFLGCPPSIVPSEKLFNT 1137
>ZBE1_HUMAN (O96006) Zinc finger BED domain containing protein 1
(dREF homolog) (Putative Ac-like transposable element)
Length = 694
Score = 59.3 bits (142), Expect = 3e-08
Identities = 111/533 (20%), Positives = 198/533 (36%), Gaps = 60/533 (11%)
Query: 159 VVEGEGFKLLCRQLQPLLTIPSRRTVARDCFQLFLDENLRLKTYFKSDCVRVALTTDCWT 218
+V+ FK+L + P +PSR+ ++ + ++ ++TD W
Sbjct: 143 IVDEPTFKVLLKTADPRYELPSRKYISTKAIPEKYGAVREVILKELAEATWCGISTDMWR 202
Query: 219 S-GQNFSYMTLTAHFIN----NDWKYEKRILSFCTVP-NHKGDTIGRKVEEILKEWGI-- 270
S QN +Y+TL AHF+ N R L VP + +TI R + E+ EWGI
Sbjct: 203 SENQNRAYVTLAAHFLGLGAPNCLSMGSRCLKTFEVPEENTAETITRVLYEVFIEWGISA 262
Query: 271 ------RNVSTITVDNASSNDVAVAYLKKRINNMGGLMGDGSFFHLRCCAHILNLVVGDG 324
N V S DVAV H+ C H N +
Sbjct: 263 KVFGATTNYGKDIVKACSLLDVAV--------------------HMPCLGHTFNAGIQQA 302
Query: 325 LKQNELS--ISSIRNSVRFVRSSPQRSAKFKECIEFARITCKKMLCLDVQTRWNTAYLML 382
+ +L +S R V + + S E + + ML + + W + ML
Sbjct: 303 FQLPKLGALLSRCRKLVEYFQQSAVAMYMLYEKQKQQNV-AHCMLVSNRVSWWGSTLAML 361
Query: 383 DGAEKFQPAFEKLEGEDSGYLEFFGEAGPPSIHDWENVRCFVRFLKIFYDATKEFSSSQ- 441
++ Q + EDS EA +W + V L+ F + S+S+
Sbjct: 362 QRLKEQQFVIAGVLVEDSNNHHLMLEAS-----EWATIEGLVELLQPFKQVAEMLSASRY 416
Query: 442 -GVSLHK-AFHQLASVHCELKRSAMNLNTVLASMGSDMKQKYNKYWGKIENINKLIYFGV 499
+S+ K H L + +K + + L+ + ++ +K + + I+ +
Sbjct: 417 PTISMVKPLLHMLLNTTLNIKETD---SKELSMAKEVIAKELSKTYQETPEIDMFLNVAT 473
Query: 500 ILDPRYKFSYVEWCFNDMYGDQPTFFTDLIAMIHTQLFKLFN-WYKDAYDQQHNSGHPSA 558
LDPRYK F ++ ++ L K+ + Y+ A D+ P
Sbjct: 474 FLDPRYKRLPFLSAF-----ERQQVENRVVEEAKGLLDKVKDGGYRPAEDKIFPV--PEE 526
Query: 559 SPSESSYVSENVIPAEVPSHLARAEAFKEHLKLKESIVKKNELERYLDEERAEDV---NF 615
P + + PA V +++ AE F + +++ ++ L +++ V N
Sbjct: 527 PPVKKLMRTSTPPPASVINNML-AEIFCQTGGVEDQEEWHAQVVEELSNFKSQKVLGLNE 585
Query: 616 DILLSWKQNSCRYPVLSSMVRDVLATPVSTVASESAFSTGGRVLDTYRSSLNP 668
D L W +P+L +++ + VA E F + V+ R+ L P
Sbjct: 586 DPLKWWSDRLALFPLLPKVLQKYWCVTATRVAPERLFGSAANVVSAKRNRLAP 638
>HOBO_DROME (P12258) Transposable element Hobo transposase
Length = 644
Score = 38.9 bits (89), Expect = 0.047
Identities = 17/54 (31%), Positives = 30/54 (55%)
Query: 599 NELERYLDEERAEDVNFDILLSWKQNSCRYPVLSSMVRDVLATPVSTVASESAF 652
+E+ERY+ + NF+++ WK N+ YP LS + +L+ P S+ + F
Sbjct: 590 DEIERYIRQRVPLSQNFEVIEWWKNNANLYPQLSKLALKLLSIPASSAELKECF 643
>FUCI_HAEIN (P44779) L-fucose isomerase (EC 5.3.1.25)
Length = 604
Score = 35.8 bits (81), Expect = 0.40
Identities = 19/75 (25%), Positives = 35/75 (46%)
Query: 553 SGHPSASPSESSYVSENVIPAEVPSHLARAEAFKEHLKLKESIVKKNELERYLDEERAED 612
+G AS SY+S + + + F+E+L ++ V E++R LD + +
Sbjct: 183 AGLAVASIRGKSYLSIGSVSMGIAGSIVNQAFFQEYLGMRNEYVDMMEIKRRLDRKIYDQ 242
Query: 613 VNFDILLSWKQNSCR 627
D+ LSW + C+
Sbjct: 243 EEVDLALSWVKQYCK 257
>DNPE_HUMAN (Q9ULA0) Aspartyl aminopeptidase (EC 3.4.11.21)
Length = 475
Score = 33.5 bits (75), Expect = 2.0
Identities = 39/141 (27%), Positives = 60/141 (41%), Gaps = 10/141 (7%)
Query: 550 QHNSGHPSASPSESSYVSENVIPAEVPSHLARAEAFKEHLKLKESIVKKNELERYLDEER 609
QH + A P +S +S ++ A P++L + E L K ++K N +RY
Sbjct: 328 QHPTAFEEAIP-KSFMISADMAHAVHPNYLDKHEENHRPLFHKGPVIKVNSKQRY----- 381
Query: 610 AEDVNFDILLSWKQNSCRYPVLSSMVRDVLATPV-STVASESAFSTGGRVLDTYRSSLNP 668
A + + L+ N + P+ MVR+ TP +T+ A G RVLD L
Sbjct: 382 ASNAVSEALIREVANKVKVPLQDLMVRN--DTPCGTTIGPILASRLGLRVLDLGSPQLAM 439
Query: 669 QMAEALICAQNWLKPTLNQFK 689
+ C L+ TL FK
Sbjct: 440 HSIREMACTTGVLQ-TLTLFK 459
>TAF4_HUMAN (O00268) Transcription initiation factor TFIID subunit 4
(Transcription initiation factor TFIID 135 kDa subunit)
(TAF(II)135) (TAFII-135) (TAFII135) (TAFII-130)
(TAFII130)
Length = 1083
Score = 32.7 bits (73), Expect = 3.4
Identities = 24/69 (34%), Positives = 29/69 (41%), Gaps = 2/69 (2%)
Query: 12 LSSTLTSGSANVAEPTEQVAVATQVPVVGLPPLPCLAKRRKPNAGGPRRTSPAWDHFIKL 71
L + + SGS A A A P G P P A R +P GGP+R P +
Sbjct: 58 LGNHVVSGSPAGAAGAGPAAPAEGAP--GAAPEPPPAGRARPGGGGPQRPGPPSPRRPLV 115
Query: 72 PDEPEPTAA 80
P P P AA
Sbjct: 116 PAGPAPPAA 124
>Y3I0_PSEAE (Q51470) Hypothetical UPF0004 protein PA3980
Length = 446
Score = 32.3 bits (72), Expect = 4.4
Identities = 28/118 (23%), Positives = 52/118 (43%), Gaps = 16/118 (13%)
Query: 245 SFCTVPNHKGDTIGRKVEEILKEWGIRNVSTITVDNASSNDVAVAYLKKRINNMGGLMGD 304
SFC VP +G+ + R ++++ E + A + V L + +N GL D
Sbjct: 162 SFCVVPYTRGEEVSRPFDDVIAE---------VIHLAENGVREVTLLGQNVNGFRGLTHD 212
Query: 305 GSFFHLRCCAHILNLVVG-DGLKQNELSISSIRNSVRFVRSSPQRSAKFKECIEFARI 361
G L A +L +V DG+++ + S + + F + Q A+ E ++F +
Sbjct: 213 G---RLADFAELLRVVAAVDGIERIRYTTS---HPLEFSDALIQAHAEVPELVKFIHL 264
>MURC_PROMP (Q7V3P8) UDP-N-acetylmuramate--L-alanine ligase (EC
6.3.2.8) (UDP-N-acetylmuramoyl-L-alanine synthetase)
Length = 473
Score = 32.0 bits (71), Expect = 5.8
Identities = 28/114 (24%), Positives = 50/114 (43%), Gaps = 18/114 (15%)
Query: 193 LDENLRLKTYFKSDCVRVALTTDCWTSGQNFSYMTLTAHFINNDWKY-EKRILSFCTVPN 251
+DE L F S+C ++ + DC + NF+ N W E +++ +PN
Sbjct: 203 IDEVLSSFKKFASNCQKLLINYDCKFTKNNFT--------SKNQWSIKESNNIAYSLIPN 254
Query: 252 --HKGDTIGRKVEE-------ILKEWGIRNVSTITVDNASSNDVAVAYLKKRIN 296
+K T+G+ E + G+ N+S IT A+ V V++ + + N
Sbjct: 255 IINKDKTVGKYYEHGKFIDIINIPVPGLHNLSNITAAIAACRMVGVSFKEIKKN 308
>METE_PYRHO (O58816) Probable methylcobalamin:homocysteine
methyltransferase (EC 2.1.1.-) (Methionine synthase)
Length = 338
Score = 31.6 bits (70), Expect = 7.5
Identities = 13/35 (37%), Positives = 21/35 (59%)
Query: 507 FSYVEWCFNDMYGDQPTFFTDLIAMIHTQLFKLFN 541
++ EW FN+ Y D+ +F DL +I+ +L L N
Sbjct: 135 YTIAEWSFNEYYPDKESFIMDLAKIINKELKMLEN 169
>DNAA_MYCAV (P49990) Chromosomal replication initiator protein dnaA
Length = 508
Score = 31.6 bits (70), Expect = 7.5
Identities = 29/112 (25%), Positives = 48/112 (41%), Gaps = 10/112 (8%)
Query: 4 VMESISQNLSSTLTSGSANVAEPTEQVAVATQVPVVGLPPLPC-LAKRRKPNA------G 56
+ +++S+ L + G +A P + V A P L+ RR+ ++ G
Sbjct: 81 ITDALSRRLGQQIQLG-VRIAPPPDDVEDALIPSAEPFPDTDADLSARRRTDSRASGERG 139
Query: 57 GPRRTSPAW-DHFIKLPDEPEPT-AACIHCHKRYLCDPKTHGTSNLLAHSKV 106
T P W ++F + P +P AA ++RY D G SN AH+ V
Sbjct: 140 AVTNTQPGWTNYFTERPHAIDPAVAAGTSLNRRYTFDTFVIGASNRFAHAAV 191
>BFPB_ECO11 (Q9S142) Outer membrane lipoprotein bfpB precursor
(Bundle-forming pilus B)
Length = 552
Score = 31.2 bits (69), Expect = 9.8
Identities = 28/100 (28%), Positives = 47/100 (47%), Gaps = 8/100 (8%)
Query: 613 VNFDILLSWKQNSCRYPVLSSMVRDVLATPVS------TVASESAFSTGGRVLDTYRSSL 666
+ FD +LS Q+S P++ +DV++ VS TVA + +TGG+ D + L
Sbjct: 96 LGFDEVLSMIQDSSGIPIVKHTTKDVISGGVSSKSLAATVAEKMNSATGGKSTDQFDHLL 155
Query: 667 NPQMAEALICAQNWLKPTLNQFKDLNINEEFELSTTVVSG 706
+E + N+ + L+ F D + + L T SG
Sbjct: 156 LEVSSEHQLMDVNY-QGALSTFLD-KVAANYNLYWTYESG 193
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.134 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 84,002,148
Number of Sequences: 164201
Number of extensions: 3663773
Number of successful extensions: 9088
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 9062
Number of HSP's gapped (non-prelim): 17
length of query: 707
length of database: 59,974,054
effective HSP length: 117
effective length of query: 590
effective length of database: 40,762,537
effective search space: 24049896830
effective search space used: 24049896830
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147434.12


