

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.3 + phase: 0 /pseudo
(659 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
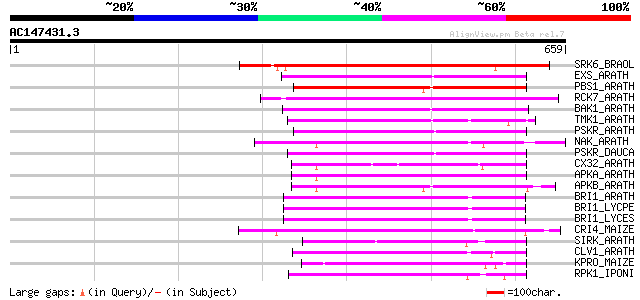
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 367 e-101
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 223 1e-57
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 221 4e-57
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 219 2e-56
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 217 9e-56
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 212 2e-54
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 209 1e-53
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 207 5e-53
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 205 3e-52
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 202 3e-51
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 198 3e-50
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 197 1e-49
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 195 3e-49
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 194 6e-49
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 194 8e-49
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 186 2e-46
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 178 4e-44
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 177 6e-44
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 174 7e-43
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 174 9e-43
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 367 bits (943), Expect = e-101
Identities = 195/391 (49%), Positives = 256/391 (64%), Gaps = 25/391 (6%)
Query: 273 IIFVSITVAVALLSCWVYSYWRKNRLSKGGMLSRTITPISFRNQ--------VQRQDSFN 324
II +++ V+V LL + RK + +K +S T RNQ + + F+
Sbjct: 447 IISLTVGVSVLLLLIMFCLWKRKQKRAKASAISIANTQ---RNQNLPMNEMVLSSKREFS 503
Query: 325 GE-------LPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSS 377
GE LP I + + ++T++FS KLG+GGFG VYKG L DG+E+AVKRLS+TS
Sbjct: 504 GEYKFEELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSV 563
Query: 378 QGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLD 437
QG++EF NEV IA+LQH NL ++LG CIEGDEK+L+YEY+ N SLD +LF + + L+
Sbjct: 564 QGTDEFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLN 623
Query: 438 WKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQC 497
W R I NG+ARGLLYLH+DSR R+IHRDLK SN+LLD M PKISDFG+AR F++D+
Sbjct: 624 WNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDET 683
Query: 498 QTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLL 557
+ T +V GTYGYM+PEYAM G+FS KSDVFSFGV+VLEI+ GK+N F+ ++ LL
Sbjct: 684 EANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLS 743
Query: 558 YTWKLWCEGKCLELIDPF-------HQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVV 610
Y W W EG+ LE++DP + EVLKCI IGLLCVQE A RP MS+VV
Sbjct: 744 YVWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
Query: 611 RMLGSDTVDLPKPTQPAFSVGRKSKNEDQIS 641
M GS+ ++P+P P + V R D S
Sbjct: 804 WMFGSEATEIPQPKPPGYCVRRSPYELDPSS 834
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 223 bits (569), Expect = 9e-58
Identities = 123/293 (41%), Positives = 176/293 (59%), Gaps = 3/293 (1%)
Query: 323 FNGELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEE 382
F L + L I ++TD FS+ +G+GGFG VYK LP + VAVK+LSE +QG+ E
Sbjct: 898 FEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNRE 957
Query: 383 FKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEE-KHKHLDWKLR 441
F E+ + K++H NL LLGYC +EK+LVYEYM N SLD L N+ + LDW R
Sbjct: 958 FMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKR 1017
Query: 442 LSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKT 501
L I G ARGL +LH +IHRD+KASN+LLD + PK++DFGLAR + T
Sbjct: 1018 LKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVST 1077
Query: 502 KRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR-NGDFFLSEHMQSLLLYTW 560
+ GT+GY+ PEY + + K DV+SFGV++LE++ GK G F +L+ +
Sbjct: 1078 V-IAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGGNLVGWAI 1136
Query: 561 KLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
+ +GK +++IDP +++ L+ + I +LC+ E A RP M V++ L
Sbjct: 1137 QKINQGKAVDVIDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 221 bits (564), Expect = 4e-57
Identities = 125/282 (44%), Positives = 178/282 (62%), Gaps = 9/282 (3%)
Query: 338 STDDFSESYKLGEGGFGPVYKGTLPD-GREVAVKRLSETSSQGSEEFKNEVIFIAKLQHR 396
+T +F LGEGGFG VYKG L G+ VAVK+L QG+ EF EV+ ++ L H
Sbjct: 82 ATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVLMLSLLHHP 141
Query: 397 NLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKH-LDWKLRLSIINGIARGLLYL 455
NL L+GYC +GD+++LVYE+MP SL+ HL + K LDW +R+ I G A+GL +L
Sbjct: 142 NLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAGAAKGLEFL 201
Query: 456 HEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLAR---TFDKDQCQTKTKRVFGTYGYMA 512
H+ + VI+RD K+SN+LLD+ +PK+SDFGLA+ T DK T RV GTYGY A
Sbjct: 202 HDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVST---RVMGTYGYCA 258
Query: 513 PEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCE-GKCLEL 571
PEYAM G +VKSDV+SFGV+ LE+I G++ D + Q+L+ + L+ + K ++L
Sbjct: 259 PEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRRKFIKL 318
Query: 572 IDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
DP + + + + + + +C+QE AA RP ++ VV L
Sbjct: 319 ADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 219 bits (557), Expect = 2e-56
Identities = 133/358 (37%), Positives = 198/358 (55%), Gaps = 9/358 (2%)
Query: 298 LSKGGMLSRTITPISFRNQVQRQDSFNGELPTIPLTIIEQSTDDFSESYKLGEGGFGPVY 357
+ K LS + ++ +QV + + T + ++T +F LGEGGFG V+
Sbjct: 64 VGKEDQLSLDVKGLNLNDQVTGKKA-----QTFTFQELAEATGNFRSDCFLGEGGFGKVF 118
Query: 358 KGTLPD-GREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYE 416
KGT+ + VA+K+L QG EF EV+ ++ H NL KL+G+C EGD+++LVYE
Sbjct: 119 KGTIEKLDQVVAIKQLDRNGVQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYE 178
Query: 417 YMPNSSLDFHLFN-EEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRDLKASNVLL 475
YMP SL+ HL K LDW R+ I G ARGL YLH+ VI+RDLK SN+LL
Sbjct: 179 YMPQGSLEDHLHVLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILL 238
Query: 476 DDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVL 535
++ PK+SDFGLA+ + RV GTYGY AP+YAM G + KSD++SFGV++L
Sbjct: 239 GEDYQPKLSDFGLAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLL 298
Query: 536 EIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCL-ELIDPFHQKTYIESEVLKCIHIGLL 594
E+I G++ D + Q+L+ + L+ + + +++DP Q Y + + + I +
Sbjct: 299 ELITGRKAIDNTKTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAM 358
Query: 595 CVQEDAADRPTMSTVVRMLGSDTVDLPKPTQPAFSVGRK-SKNEDQISKNSKDNSVDE 651
CVQE RP +S VV L P P+ S G+ S + D+ + + + V E
Sbjct: 359 CVQEQPTMRPVVSDVVLALNFLASSKYDPNSPSSSSGKNPSFHRDRDDEEKRPHLVKE 416
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 217 bits (552), Expect = 9e-56
Identities = 129/296 (43%), Positives = 170/296 (56%), Gaps = 5/296 (1%)
Query: 325 GELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSE-EF 383
G+L L ++ ++D+FS LG GGFG VYKG L DG VAVKRL E +QG E +F
Sbjct: 272 GQLKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQF 331
Query: 384 KNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNE-EKHKHLDWKLRL 442
+ EV I+ HRNL +L G+C+ E++LVY YM N S+ L E LDW R
Sbjct: 332 QTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQ 391
Query: 443 SIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTK 502
I G ARGL YLH+ ++IHRD+KA+N+LLD+E + DFGLA+ D T
Sbjct: 392 RIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHV-TT 450
Query: 503 RVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW-- 560
V GT G++APEY G S K+DVF +GV++LE+I G+R D + ++L W
Sbjct: 451 AVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVK 510
Query: 561 KLWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSD 616
L E K L+D Q Y + EV + I + LLC Q +RP MS VVRML D
Sbjct: 511 GLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLEGD 566
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 212 bits (540), Expect = 2e-54
Identities = 124/304 (40%), Positives = 183/304 (59%), Gaps = 13/304 (4%)
Query: 330 IPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSE--TSSQGSEEFKNEV 387
I + ++ T++FS LG GGFG VYKG L DG ++AVKR+ + +G EFK+E+
Sbjct: 576 ISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEI 635
Query: 388 IFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLF--NEEKHKHLDWKLRLSII 445
+ K++HR+L LLGYC++G+EK+LVYEYMP +L HLF +EE K L WK RL++
Sbjct: 636 AVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLA 695
Query: 446 NGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVF 505
+ARG+ YLH + IHRDLK SN+LL D+M K++DFGL R + + +T R+
Sbjct: 696 LDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIET-RIA 754
Query: 506 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW-KLWC 564
GT+GY+APEYA+ G + K DV+SFGV+++E+I G+++ D S+ +S+ L +W K
Sbjct: 755 GTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLD--ESQPEESIHLVSWFKRMY 812
Query: 565 EGKCLELIDPFHQKTYIESEVLKCIH----IGLLCVQEDAADRPTMSTVVRMLGSDTVDL 620
K ++ E L +H + C + RP M V +L S V+L
Sbjct: 813 INKEASFKKAIDTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNIL-SSLVEL 871
Query: 621 PKPT 624
KP+
Sbjct: 872 WKPS 875
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 209 bits (533), Expect = 1e-53
Identities = 111/277 (40%), Positives = 163/277 (58%), Gaps = 2/277 (0%)
Query: 338 STDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRN 397
ST+ F ++ +G GGFG VYK TLPDG++VA+K+LS Q EF+ EV +++ QH N
Sbjct: 730 STNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRAQHPN 789
Query: 398 LAKLLGYCIEGDEKILVYEYMPNSSLDFHLF-NEEKHKHLDWKLRLSIINGIARGLLYLH 456
L L G+C ++++L+Y YM N SLD+ L + L WK RL I G A+GLLYLH
Sbjct: 790 LVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLYLH 849
Query: 457 EDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYA 516
E ++HRD+K+SN+LLD+ N ++DFGLAR + T V GT GY+ PEY
Sbjct: 850 EGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTDLV-GTLGYIPPEYG 908
Query: 517 MAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFH 576
A + + K DV+SFGV++LE++ KR D + + L+ + K+ E + E+ DP
Sbjct: 909 QASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASEVFDPLI 968
Query: 577 QKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
+ E+ + + I LC+ E+ RPT +V L
Sbjct: 969 YSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 207 bits (528), Expect = 5e-53
Identities = 132/384 (34%), Positives = 205/384 (53%), Gaps = 30/384 (7%)
Query: 291 SYWRKNR-LSKGGMLSRTITPISFRNQVQRQDSFNGELPTIPLTIIEQSTDDFSESYKLG 349
S W ++ LS+ G + S+ + + + N L L+ ++ +T +F +G
Sbjct: 16 STWLSSKFLSRDGSKGSSTASFSYMPRTEGEILQNANLKNFSLSELKSATRNFRPDSVVG 75
Query: 350 EGGFGPVYKGTLPD----------GREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLA 399
EGGFG V+KG + + G +AVKRL++ QG E+ E+ ++ +L H NL
Sbjct: 76 EGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQEGFQGHREWLAEINYLGQLDHPNLV 135
Query: 400 KLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEK-HKHLDWKLRLSIINGIARGLLYLHED 458
KL+GYC+E + ++LVYE+M SL+ HLF ++ L W R+ + G ARGL +LH +
Sbjct: 136 KLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQPLSWNTRVRMALGAARGLAFLH-N 194
Query: 459 SRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMA 518
++ +VI+RD KASN+LLD N K+SDFGLAR + RV GT GY APEY
Sbjct: 195 AQPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPMGDNSHVSTRVMGTQGYAAPEYLAT 254
Query: 519 GLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWK---LWCEGKCLELIDPF 575
G SVKSDV+SFGV++LE++ G+R D ++ + L W L + + L ++DP
Sbjct: 255 GHLSVKSDVYSFGVVLLELLSGRRAID--KNQPVGEHNLVDWARPYLTNKRRLLRVMDPR 312
Query: 576 HQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRMLGSDTVDLPKPTQPAFSVGRKSK 635
Q Y + LK + L C+ DA RPTM+ +V+ T + +++
Sbjct: 313 LQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVK------------TMEELHIQKEAS 360
Query: 636 NEDQISKNSKDNSVDEETITIVSP 659
E Q + S DN +++ + P
Sbjct: 361 KEQQNPQISIDNIINKSPQAVNYP 384
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 205 bits (522), Expect = 3e-52
Identities = 117/286 (40%), Positives = 168/286 (57%), Gaps = 4/286 (1%)
Query: 330 IPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIF 389
+ L I +ST F+++ +G GGFG VYK TLPDG +VA+KRLS + Q EF+ EV
Sbjct: 731 LSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVET 790
Query: 390 IAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNE-EKHKHLDWKLRLSIINGI 448
+++ QH NL LLGYC ++K+L+Y YM N SLD+ L + + LDWK RL I G
Sbjct: 791 LSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGA 850
Query: 449 ARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTY 508
A GL YLH+ ++HRD+K+SN+LL D ++DFGLAR T V GT
Sbjct: 851 AEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTDLV-GTL 909
Query: 509 GYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKC 568
GY+ PEY A + + K DV+SFGV++LE++ G+R D + L+ + ++ E +
Sbjct: 910 GYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRE 969
Query: 569 LELIDPF-HQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
E+ DPF + K + E E+L + I C+ E+ RPT +V L
Sbjct: 970 SEIFDPFIYDKDHAE-EMLLVLEIACRCLGENPKTRPTTQQLVSWL 1014
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 202 bits (513), Expect = 3e-51
Identities = 120/292 (41%), Positives = 167/292 (57%), Gaps = 18/292 (6%)
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLP----------DGREVAVKRLSETSSQGSEEFK 384
++ +T +F LG+GGFG VY+G + G VA+KRL+ S QG E++
Sbjct: 79 LKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGFAEWR 138
Query: 385 NEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSI 444
+EV F+ L HRNL KLLGYC E E +LVYE+MP SL+ HLF ++ W LR+ I
Sbjct: 139 SEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFR--RNDPFPWDLRIKI 196
Query: 445 INGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRV 504
+ G ARGL +LH R VI+RD KASN+LLD + K+SDFGLA+ D+ T R+
Sbjct: 197 VIGAARGLAFLHSLQR-EVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTRI 255
Query: 505 FGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW---K 561
GTYGY APEY G VKSDVF+FGV++LEI+ G + +SL+ W +
Sbjct: 256 MGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRGQESLV--DWLRPE 313
Query: 562 LWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
L + + +++D + Y + I L C++ D +RP M VV +L
Sbjct: 314 LSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVL 365
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 198 bits (504), Expect = 3e-50
Identities = 117/291 (40%), Positives = 168/291 (57%), Gaps = 13/291 (4%)
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPD----------GREVAVKRLSETSSQGSEEFK 384
++ +T +F LGEGGFG V+KG + + G +AVK+L++ QG +E+
Sbjct: 61 LKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQGHQEWL 120
Query: 385 NEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKH-KHLDWKLRLS 443
EV ++ + HR+L KL+GYC+E + ++LVYE+MP SL+ HLF + + L WKLRL
Sbjct: 121 AEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSWKLRLK 180
Query: 444 IINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKR 503
+ G A+GL +LH S RVI+RD K SN+LLD E N K+SDFGLA+ + R
Sbjct: 181 VALGAAKGLAFLH-SSETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKSHVSTR 239
Query: 504 VFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWK-L 562
V GT+GY APEY G + KSDV+SFGV++LE++ G+R D ++L+ + L
Sbjct: 240 VMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEWAKPYL 299
Query: 563 WCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
+ K +ID Q Y E K + L C+ + RP MS VV L
Sbjct: 300 VNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHL 350
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 197 bits (500), Expect = 1e-49
Identities = 124/338 (36%), Positives = 182/338 (53%), Gaps = 36/338 (10%)
Query: 335 IEQSTDDFSESYKLGEGGFGPVYKGTLPD----------GREVAVKRLSETSSQGSEEFK 384
++ +T +F LGEGGFG V+KG + + G +AVK+L++ QG +E+
Sbjct: 62 LKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQGHQEWL 121
Query: 385 NEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKH-KHLDWKLRLS 443
EV ++ + H NL KL+GYC+E + ++LVYE+MP SL+ HLF + + L W LRL
Sbjct: 122 AEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSWTLRLK 181
Query: 444 IINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLAR---TFDKDQCQTK 500
+ G A+GL +LH ++ VI+RD K SN+LLD E N K+SDFGLA+ T DK T
Sbjct: 182 VALGAAKGLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKSHVST- 239
Query: 501 TKRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW 560
R+ GTYGY APEY G + KSDV+S+GV++LE++ G+R D Q L+ +
Sbjct: 240 --RIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEWAR 297
Query: 561 KLWC-EGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML------ 613
L + K +ID Q Y E K + L C+ + RP M+ VV L
Sbjct: 298 PLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLEHIQTL 357
Query: 614 ---GSDTVDLPKPTQPAFSVGRKSKNEDQISKNSKDNS 648
G +D+ + R + D ++ N K N+
Sbjct: 358 NEAGGRNIDMVQ--------RRMRRRSDSVAINQKPNA 387
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 195 bits (496), Expect = 3e-49
Identities = 116/293 (39%), Positives = 165/293 (55%), Gaps = 9/293 (3%)
Query: 326 ELPTIPLTIIE--QSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEF 383
E P LT + Q+T+ F +G GGFG VYK L DG VA+K+L S QG EF
Sbjct: 865 EKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREF 924
Query: 384 KNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHK-HLDWKLRL 442
E+ I K++HRNL LLGYC GDE++LVYE+M SL+ L + +K L+W R
Sbjct: 925 MAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRR 984
Query: 443 SIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTK 502
I G ARGL +LH + +IHRD+K+SNVLLD+ + ++SDFG+AR
Sbjct: 985 KIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 1044
Query: 503 RVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW-K 561
+ GT GY+ PEY + S K DV+S+GV++LE++ GKR D S L W K
Sbjct: 1045 TLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD---SPDFGDNNLVGWVK 1101
Query: 562 LWCEGKCLELIDP--FHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRM 612
+ + ++ DP + +E E+L+ + + + C+ + A RPTM V+ M
Sbjct: 1102 QHAKLRISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAM 1154
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 194 bits (493), Expect = 6e-49
Identities = 116/293 (39%), Positives = 164/293 (55%), Gaps = 9/293 (3%)
Query: 326 ELPTIPLTIIE--QSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEF 383
E P LT + ++T+ F +G GGFG VYK L DG VA+K+L S QG EF
Sbjct: 870 EKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREF 929
Query: 384 KNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHK-HLDWKLRL 442
E+ I K++HRNL LLGYC G+E++LVYEYM SL+ L + +K L+W R
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARR 989
Query: 443 SIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTK 502
I G ARGL +LH + +IHRD+K+SNVLLD+ + ++SDFG+AR
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 1049
Query: 503 RVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW-K 561
+ GT GY+ PEY + S K DV+S+GV++LE++ GK+ D S L W K
Sbjct: 1050 TLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD---SADFGDNNLVGWVK 1106
Query: 562 LWCEGKCLELIDP--FHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRM 612
L +GK ++ D + IE E+L+ + + C+ + RPTM V+ M
Sbjct: 1107 LHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 194 bits (492), Expect = 8e-49
Identities = 116/293 (39%), Positives = 164/293 (55%), Gaps = 9/293 (3%)
Query: 326 ELPTIPLTIIE--QSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEF 383
E P LT + ++T+ F +G GGFG VYK L DG VA+K+L S QG EF
Sbjct: 870 EKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREF 929
Query: 384 KNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHK-HLDWKLRL 442
E+ I K++HRNL LLGYC G+E++LVYEYM SL+ L + +K L+W R
Sbjct: 930 TAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARR 989
Query: 443 SIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTK 502
I G ARGL +LH + +IHRD+K+SNVLLD+ + ++SDFG+AR
Sbjct: 990 KIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVS 1049
Query: 503 RVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTW-K 561
+ GT GY+ PEY + S K DV+S+GV++LE++ GK+ D S L W K
Sbjct: 1050 TLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD---SADFGDNNLVGWVK 1106
Query: 562 LWCEGKCLELIDP--FHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRM 612
L +GK ++ D + IE E+L+ + + C+ + RPTM V+ M
Sbjct: 1107 LHAKGKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 186 bits (472), Expect = 2e-46
Identities = 129/402 (32%), Positives = 207/402 (51%), Gaps = 25/402 (6%)
Query: 272 IIIFVSITVAVALLSC-WVYSYWRKNRLSKGGMLSRTITPISFRN-----QVQRQDSFNG 325
I+ V + ++V++ +C +V R + S + T SFR Q +D
Sbjct: 429 IVFAVVLVLSVSVTTCLYVRHKLRHCQCSNRELRLAKSTAYSFRKDNMKIQPDMEDLKIR 488
Query: 326 ELPTIPLTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSS--QGSEEF 383
+EQ+T FSE ++G+G F V+KG L DG VAVKR + S + S+EF
Sbjct: 489 RAQEFSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEF 548
Query: 384 KNEVIFIAKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKH--KHLDWKLR 441
NE+ +++L H +L LLGYC +G E++LVYE+M + SL HL ++ + K L+W R
Sbjct: 549 HNELDLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARR 608
Query: 442 LSIINGIARGLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKT 501
++I ARG+ YLH + VIHRD+K+SN+L+D++ N +++DFGL+ D +
Sbjct: 609 VTIAVQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLS 668
Query: 502 KRVFGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWK 561
+ GT GY+ PEY + KSDV+SFGV++LEI+ G++ D E +++ +
Sbjct: 669 ELPAGTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEE--GNIVEWAVP 726
Query: 562 LWCEGKCLELIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVR--------ML 613
L G ++DP + K + CV+ DRP+M V ++
Sbjct: 727 LIKAGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALEHALALLM 786
Query: 614 GSDTVDLP-KPTQPAFSVGRKSKNEDQISKNSKDNSVDEETI 654
GS ++ P PT+ R K +S+ S ++S E +
Sbjct: 787 GSPCIEQPILPTEVVLGSSRMHK----VSQMSSNHSCSENEL 824
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 178 bits (452), Expect = 4e-44
Identities = 102/272 (37%), Positives = 158/272 (57%), Gaps = 15/272 (5%)
Query: 348 LGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCIE 407
+G+GGFG VY G + +G +VAVK LSE S+QG +EF+ EV + ++ H NL L+GYC E
Sbjct: 580 IGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFRAEVDLLMRVHHTNLTSLVGYCNE 638
Query: 408 GDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHRD 467
+ +L+YEYM N +L +L + L W+ RL I A+GL YLH + ++HRD
Sbjct: 639 INHMVLIYEYMANENLGDYLAGKRSFI-LSWEERLKISLDAAQGLEYLHNGCKPPIVHRD 697
Query: 468 LKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSDV 527
+K +N+LL++++ K++DFGL+R+F + + V G+ GY+ PEY + KSDV
Sbjct: 698 VKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSIGYLDPEYYSTRQMNEKSDV 757
Query: 528 FSFGVLVLEIIYGK------RNGDFFLSEHMQSLLLYTWKLWCEGKCLELIDPFHQKTYI 581
+S GV++LE+I G+ + +S+H++S+L G ++D ++ Y
Sbjct: 758 YSLGVVLLEVITGQPAIASSKTEKVHISDHVRSIL-------ANGDIRGIVDQRLRERYD 810
Query: 582 ESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
K I L C + +A RPTMS VV L
Sbjct: 811 VGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 177 bits (450), Expect = 6e-44
Identities = 103/288 (35%), Positives = 160/288 (54%), Gaps = 16/288 (5%)
Query: 336 EQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEE-FKNEVIFIAKLQ 394
E + E +G+GG G VY+G++P+ +VA+KRL + S+ F E+ + +++
Sbjct: 686 EDVLECLKEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIR 745
Query: 395 HRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLY 454
HR++ +LLGY D +L+YEYMPN SL L + K HL W+ R + A+GL Y
Sbjct: 746 HRHIVRLLGYVANKDTNLLLYEYMPNGSLG-ELLHGSKGGHLQWETRHRVAVEAAKGLCY 804
Query: 455 LHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPE 514
LH D ++HRD+K++N+LLD + ++DFGLA+ + G+YGY+APE
Sbjct: 805 LHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPE 864
Query: 515 YAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHMQSLLLYTWKLWCEGKCLE---- 570
YA KSDV+SFGV++LE+I GK+ + E + + + W E + +
Sbjct: 865 YAYTLKVDEKSDVYSFGVVLLELIAGKKP----VGEFGEGVDIVRWVRNTEEEITQPSDA 920
Query: 571 -----LIDPFHQKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
++DP Y + V+ I ++CV+E+AA RPTM VV ML
Sbjct: 921 AIVVAIVDP-RLTGYPLTSVIHVFKIAMMCVEEEAAARPTMREVVHML 967
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 174 bits (441), Expect = 7e-43
Identities = 104/277 (37%), Positives = 157/277 (56%), Gaps = 13/277 (4%)
Query: 347 KLGEGGFGPVYKGTLPDGREVAVKRLSETSSQGSEEFKNEVIFIAKLQHRNLAKLLGYCI 406
+LG G G VYKG L D R VAVK+L E QG E F+ E+ I ++ H NL ++ G+C
Sbjct: 539 ELGRGESGTVYKGVLEDDRHVAVKKL-ENVRQGKEVFQAELSVIGRINHMNLVRIWGFCS 597
Query: 407 EGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIARGLLYLHEDSRLRVIHR 466
EG ++LV EY+ N SL LF+E + LDW+ R +I G+A+GL YLH + VIH
Sbjct: 598 EGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVAKGLAYLHHECLEWVIHC 657
Query: 467 DLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGYMAPEYAMAGLFSVKSD 526
D+K N+LLD PKI+DFGL + ++ V GT GY+APE+ + + K D
Sbjct: 658 DVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLGYIAPEWVSSLPITAKVD 717
Query: 527 VFSFGVLVLEIIYGKRNGDFF-LSEHMQSLLLYTWKLW---CEGKCLELIDPF------H 576
V+S+GV++LE++ G R + ++ + S+L ++ EG+ ID +
Sbjct: 718 VYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRMLSAKLEGEEQSWIDGYLDSKLNR 777
Query: 577 QKTYIESEVLKCIHIGLLCVQEDAADRPTMSTVVRML 613
Y+++ L I + + C++ED + RPTM V+ L
Sbjct: 778 PVNYVQARTL--IKLAVSCLEEDRSKRPTMEHAVQTL 812
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 174 bits (440), Expect = 9e-43
Identities = 103/293 (35%), Positives = 161/293 (54%), Gaps = 17/293 (5%)
Query: 332 LTIIEQSTDDFSESYKLGEGGFGPVYKGTLPDGREVAVKRLSETS-SQGSEEFKNEVIFI 390
L + ++T++ ++ Y +G+G G +YK TL + AVK+L T GS E+ I
Sbjct: 806 LNKVLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETI 865
Query: 391 AKLQHRNLAKLLGYCIEGDEKILVYEYMPNSSLDFHLFNEEKHKHLDWKLRLSIINGIAR 450
K++HRNL KL + + + +++Y YM N SL L K LDW R +I G A
Sbjct: 866 GKVRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAH 925
Query: 451 GLLYLHEDSRLRVIHRDLKASNVLLDDEMNPKISDFGLARTFDKDQCQTKTKRVFGTYGY 510
GL YLH D ++HRD+K N+LLD ++ P ISDFG+A+ D+ + V GT GY
Sbjct: 926 GLAYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGY 985
Query: 511 MAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR------NGDFFLSEHMQSLLLYTWKLWC 564
MAPE A + S +SDV+S+GV++LE+I K+ NG+ + ++S+ T
Sbjct: 986 MAPENAFTTVKSRESDVYSYGVVLLELITRKKALDPSFNGETDIVGWVRSVWTQT----- 1040
Query: 565 EGKCLELIDPFHQKTYIESEVL----KCIHIGLLCVQEDAADRPTMSTVVRML 613
G+ +++DP I+S V+ + + + L C +++ RPTM VV+ L
Sbjct: 1041 -GEIQKIVDPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 78,560,644
Number of Sequences: 164201
Number of extensions: 3401271
Number of successful extensions: 12284
Number of sequences better than 10.0: 1639
Number of HSP's better than 10.0 without gapping: 1127
Number of HSP's successfully gapped in prelim test: 512
Number of HSP's that attempted gapping in prelim test: 8624
Number of HSP's gapped (non-prelim): 2010
length of query: 659
length of database: 59,974,054
effective HSP length: 117
effective length of query: 542
effective length of database: 40,762,537
effective search space: 22093295054
effective search space used: 22093295054
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147431.3


