

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147431.1 + phase: 0 /pseudo
(493 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
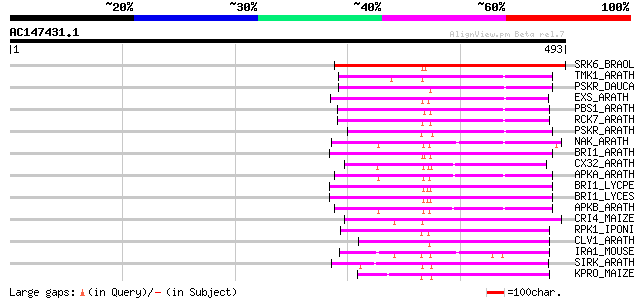
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 233 1e-60
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 162 2e-39
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 156 1e-37
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 156 1e-37
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 155 2e-37
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 155 3e-37
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 155 3e-37
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 151 4e-36
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 150 9e-36
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 149 2e-35
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 148 3e-35
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 145 2e-34
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 145 2e-34
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 144 5e-34
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 132 2e-30
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 129 2e-29
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 125 3e-28
IRA1_MOUSE (Q62406) Interleukin-1 receptor-associated kinase 1 (... 124 4e-28
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 124 7e-28
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 120 6e-27
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 233 bits (593), Expect = 1e-60
Identities = 122/231 (52%), Positives = 155/231 (66%), Gaps = 26/231 (11%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKN 348
+LP I + + ++T+NFS KLG+GGFG VYKG L DG EIA KRLS+TS QG +EF N
Sbjct: 512 ELPLIEMETVVKATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMN 571
Query: 349 EVIFIAKLQHRNLVKLL------DEE--------------------KHKHLDWKLRLSII 382
EV IA+LQH NLV++L DE+ + L+W R I
Sbjct: 572 EVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGKTRRSKLNWNERFDIT 631
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
G+ARGLLYLH+DS R+IHRDLK SN+LLD M PKISDFG+AR FE+D+ T +V+
Sbjct: 632 NGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVV 691
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNLETL 493
GTYGYM+PEYAM G+FS KSDVFSFGV+VLEI+ GK+N F+ ++ + L
Sbjct: 692 GTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLL 742
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 162 bits (409), Expect = 2e-39
Identities = 92/220 (41%), Positives = 131/220 (58%), Gaps = 31/220 (14%)
Query: 293 IPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSE--TSGQGLEEFKNEV 350
I + V++ T+NFS LG GGFG VYKG L DGT+IA KR+ +G+G EFK+E+
Sbjct: 576 ISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEI 635
Query: 351 IFIAKLQHRNLVKLL----------------------------DEEKHKHLDWKLRLSII 382
+ K++HR+LV LL EE K L WK RL++
Sbjct: 636 AVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLA 695
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVI 442
+ARG+ YLH + IHRDLK SN+LL D+M K++DFGL R + + +T R+
Sbjct: 696 LDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIET-RIA 754
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
GT+GY+APEYA+ G + K DV+SFGV+++E+I G+++ D
Sbjct: 755 GTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLD 794
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 156 bits (395), Expect = 1e-37
Identities = 89/217 (41%), Positives = 125/217 (57%), Gaps = 28/217 (12%)
Query: 293 IPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIF 352
+ L I +ST +F+++ +G GGFG VYK TLPDGT++A KRLS +GQ EF+ EV
Sbjct: 731 LSLDDILKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVET 790
Query: 353 IAKLQHRNLVKLLDEEKHKH---------------------------LDWKLRLSIIKGI 385
+++ QH NLV LL +K+ LDWK RL I +G
Sbjct: 791 LSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGA 850
Query: 386 ARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTY 445
A GL YLH+ ++HRD+K+SN+LL D ++DFGLAR H T ++GT
Sbjct: 851 AEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTD-LVGTL 909
Query: 446 GYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
GY+ PEY A + + K DV+SFGV++LE++ G+R D
Sbjct: 910 GYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMD 946
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 156 bits (394), Expect = 1e-37
Identities = 90/220 (40%), Positives = 122/220 (54%), Gaps = 28/220 (12%)
Query: 286 FNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEE 345
F L + L I ++TD+FS+ +G+GGFG VYK LP +A K+LSE QG E
Sbjct: 898 FEQPLLKVRLGDIVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNRE 957
Query: 346 FKNEVIFIAKLQHRNLVKLL-----DEEKH----------------------KHLDWKLR 378
F E+ + K++H NLV LL EEK + LDW R
Sbjct: 958 FMAEMETLGKVKHPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKR 1017
Query: 379 LSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKT 438
L I G ARGL +LH +IHRD+KASN+LLD + PK++DFGLAR + H T
Sbjct: 1018 LKIAVGAARGLAFLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVST 1077
Query: 439 KRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGK 478
+ GT+GY+ PEY + + K DV+SFGV++LE++ GK
Sbjct: 1078 V-IAGTFGYIPPEYGQSARATTKGDVYSFGVILLELVTGK 1116
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 155 bits (392), Expect = 2e-37
Identities = 94/217 (43%), Positives = 123/217 (56%), Gaps = 30/217 (13%)
Query: 292 TIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPD-GTEIAAKRLSETSGQGLEEFKNEV 350
T + +T NF LGEGGFG VYKG L G +A K+L QG EF EV
Sbjct: 73 TFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEV 132
Query: 351 IFIAKLQHRNLVKLLDE-------------------EKHKH--------LDWKLRLSIIK 383
+ ++ L H NLV L+ E H H LDW +R+ I
Sbjct: 133 LMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAA 192
Query: 384 GIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEK-DQCHTKTKRVI 442
G A+GL +LH+ + VI+RD K+SN+LLD+ +PK+SDFGLA+ D+ H T RV+
Sbjct: 193 GAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVST-RVM 251
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR 479
GTYGY APEYAM G +VKSDV+SFGV+ LE+I G++
Sbjct: 252 GTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRK 288
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 155 bits (391), Expect = 3e-37
Identities = 91/217 (41%), Positives = 123/217 (55%), Gaps = 30/217 (13%)
Query: 292 TIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAA-KRLSETSGQGLEEFKNEV 350
T + ++T NF LGEGGFG V+KGT+ ++ A K+L QG+ EF EV
Sbjct: 90 TFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGIREFVVEV 149
Query: 351 IFIAKLQHRNLVKLL-------------------DEEKHKH--------LDWKLRLSIIK 383
+ ++ H NLVKL+ E H H LDW R+ I
Sbjct: 150 LTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWNTRMKIAA 209
Query: 384 GIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEK-DQCHTKTKRVI 442
G ARGL YLH+ VI+RDLK SN+LL ++ PK+SDFGLA+ D+ H T RV+
Sbjct: 210 GAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTHVST-RVM 268
Query: 443 GTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR 479
GTYGY AP+YAM G + KSD++SFGV++LE+I G++
Sbjct: 269 GTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRK 305
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 155 bits (391), Expect = 3e-37
Identities = 88/209 (42%), Positives = 122/209 (58%), Gaps = 28/209 (13%)
Query: 301 STDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRN 360
ST++F ++ +G GGFG VYK TLPDG ++A K+LS GQ EF+ EV +++ QH N
Sbjct: 730 STNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETLSRAQHPN 789
Query: 361 LVKL---------------------LDEEKHKHLD------WKLRLSIIKGIARGLLYLH 393
LV L LD H+ D WK RL I +G A+GLLYLH
Sbjct: 790 LVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAAKGLLYLH 849
Query: 394 EDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYA 453
E ++HRD+K+SN+LLD+ N ++DFGLAR + H T ++GT GY+ PEY
Sbjct: 850 EGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTD-LVGTLGYIPPEYG 908
Query: 454 MAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
A + + K DV+SFGV++LE++ KR D
Sbjct: 909 QASVATYKGDVYSFGVVLLELLTDKRPVD 937
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 151 bits (381), Expect = 4e-36
Identities = 99/244 (40%), Positives = 133/244 (53%), Gaps = 42/244 (17%)
Query: 287 NGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPD----------GTEIAAKRLS 336
N +L L+ ++ +T NF +GEGGFG V+KG + + G IA KRL+
Sbjct: 50 NANLKNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLN 109
Query: 337 ETSGQGLEEFKNEVIFIAKLQHRNLVKLLD---EEKHK---------------------- 371
+ QG E+ E+ ++ +L H NLVKL+ EE+H+
Sbjct: 110 QEGFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTF 169
Query: 372 --HLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAF 429
L W R+ + G ARGL +LH P +VI+RD KASN+LLD N K+SDFGLAR
Sbjct: 170 YQPLSWNTRVRMALGAARGLAFLHNAQP-QVIYRDFKASNILLDSNYNAKLSDFGLARDG 228
Query: 430 EK-DQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFF--LS 486
D H T RV+GT GY APEY G SVKSDV+SFGV++LE++ G+R D +
Sbjct: 229 PMGDNSHVST-RVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVG 287
Query: 487 EHNL 490
EHNL
Sbjct: 288 EHNL 291
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 150 bits (378), Expect = 9e-36
Identities = 88/225 (39%), Positives = 120/225 (53%), Gaps = 27/225 (12%)
Query: 285 SFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLE 344
+F L + + Q+T+ F +G GGFG VYK L DG+ +A K+L SGQG
Sbjct: 863 AFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDR 922
Query: 345 EFKNEVIFIAKLQHRNLVKLL------DE-------------EKHKH--------LDWKL 377
EF E+ I K++HRNLV LL DE E H L+W
Sbjct: 923 EFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWST 982
Query: 378 RLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTK 437
R I G ARGL +LH + +IHRD+K+SNVLLD+ + ++SDFG+AR H
Sbjct: 983 RRKIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLS 1042
Query: 438 TKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
+ GT GY+ PEY + S K DV+S+GV++LE++ GKR D
Sbjct: 1043 VSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTD 1087
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 149 bits (376), Expect = 2e-35
Identities = 92/214 (42%), Positives = 120/214 (55%), Gaps = 35/214 (16%)
Query: 298 IQQSTDNFSESFKLGEGGFGPVYKGTLP----------DGTEIAAKRLSETSGQGLEEFK 347
++ +T NF LG+GGFG VY+G + G +A KRL+ S QG E++
Sbjct: 79 LKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESVQGFAEWR 138
Query: 348 NEVIFIAKLQHRNLVKLLD---EEKH--------------KHL-------DWKLRLSIIK 383
+EV F+ L HRNLVKLL E+K HL W LR+ I+
Sbjct: 139 SEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHLFRRNDPFPWDLRIKIVI 198
Query: 384 GIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIG 443
G ARGL +LH VI+RD KASN+LLD + K+SDFGLA+ D+ T R++G
Sbjct: 199 GAARGLAFLHS-LQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADEKSHVTTRIMG 257
Query: 444 TYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYG 477
TYGY APEY G VKSDVF+FGV++LEI+ G
Sbjct: 258 TYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTG 291
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 148 bits (374), Expect = 3e-35
Identities = 92/232 (39%), Positives = 130/232 (55%), Gaps = 40/232 (17%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPD----------GTEIAAKRLSET 338
+L + ++ +T NF LGEGGFG V+KG + + G IA K+L++
Sbjct: 52 NLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQD 111
Query: 339 SGQGLEEFKNEVIFIAKLQHRNLVKLLD---EEKHK------------------------ 371
QG +E+ EV ++ + HR+LVKL+ E++H+
Sbjct: 112 GWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQ 171
Query: 372 HLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFE- 430
L WKLRL + G A+GL +LH S RVI+RD K SN+LLD E N K+SDFGLA+
Sbjct: 172 PLSWKLRLKVALGAAKGLAFLHS-SETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPI 230
Query: 431 KDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
D+ H T RV+GT+GY APEY G + KSDV+SFGV++LE++ G+R D
Sbjct: 231 GDKSHVST-RVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVD 281
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 145 bits (367), Expect = 2e-34
Identities = 85/225 (37%), Positives = 120/225 (52%), Gaps = 27/225 (12%)
Query: 285 SFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLE 344
+F L + + ++T+ F +G GGFG VYK L DG+ +A K+L SGQG
Sbjct: 868 AFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDR 927
Query: 345 EFKNEVIFIAKLQHRNLVKLLD-----EEK----------------HKH------LDWKL 377
EF E+ I K++HRNLV LL EE+ H L+W
Sbjct: 928 EFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPA 987
Query: 378 RLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTK 437
R I G ARGL +LH + +IHRD+K+SNVLLD+ + ++SDFG+AR H
Sbjct: 988 RRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLS 1047
Query: 438 TKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
+ GT GY+ PEY + S K DV+S+GV++LE++ GK+ D
Sbjct: 1048 VSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD 1092
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 145 bits (367), Expect = 2e-34
Identities = 85/225 (37%), Positives = 120/225 (52%), Gaps = 27/225 (12%)
Query: 285 SFNGDLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLE 344
+F L + + ++T+ F +G GGFG VYK L DG+ +A K+L SGQG
Sbjct: 868 AFEKPLRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDR 927
Query: 345 EFKNEVIFIAKLQHRNLVKLLD-----EEK----------------HKH------LDWKL 377
EF E+ I K++HRNLV LL EE+ H L+W
Sbjct: 928 EFTAEMETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPA 987
Query: 378 RLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTK 437
R I G ARGL +LH + +IHRD+K+SNVLLD+ + ++SDFG+AR H
Sbjct: 988 RRKIAIGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLS 1047
Query: 438 TKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
+ GT GY+ PEY + S K DV+S+GV++LE++ GK+ D
Sbjct: 1048 VSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTD 1092
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 144 bits (363), Expect = 5e-34
Identities = 88/232 (37%), Positives = 128/232 (54%), Gaps = 40/232 (17%)
Query: 289 DLPTIPLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPD----------GTEIAAKRLSET 338
+L + ++ +T NF LGEGGFG V+KG + + G IA K+L++
Sbjct: 53 NLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQD 112
Query: 339 SGQGLEEFKNEVIFIAKLQHRNLVKLLD---EEKHK------------------------ 371
QG +E+ EV ++ + H NLVKL+ E++H+
Sbjct: 113 GWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQ 172
Query: 372 HLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEK 431
L W LRL + G A+GL +LH ++ VI+RD K SN+LLD E N K+SDFGLA+
Sbjct: 173 PLSWTLRLKVALGAAKGLAFLH-NAETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPT 231
Query: 432 -DQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGD 482
D+ H T R++GTYGY APEY G + KSDV+S+GV++LE++ G+R D
Sbjct: 232 GDKSHVST-RIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVD 282
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 132 bits (333), Expect = 2e-30
Identities = 78/223 (34%), Positives = 121/223 (53%), Gaps = 30/223 (13%)
Query: 298 IQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSG--QGLEEFKNEVIFIAK 355
++Q+T FSE ++G+G F V+KG L DGT +A KR + S + +EF NE+ +++
Sbjct: 498 LEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELDLLSR 557
Query: 356 LQHRNLVKLL----------------------------DEEKHKHLDWKLRLSIIKGIAR 387
L H +L+ LL D K L+W R++I AR
Sbjct: 558 LNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIAVQAAR 617
Query: 388 GLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGY 447
G+ YLH + VIHRD+K+SN+L+D++ N +++DFGL+ D ++ GT GY
Sbjct: 618 GIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPAGTLGY 677
Query: 448 MAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKRNGDFFLSEHNL 490
+ PEY + KSDV+SFGV++LEI+ G++ D E N+
Sbjct: 678 LDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEGNI 720
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 129 bits (323), Expect = 2e-29
Identities = 76/212 (35%), Positives = 112/212 (51%), Gaps = 27/212 (12%)
Query: 295 LTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETS-GQGLEEFKNEVIFI 353
L + ++T+N ++ + +G+G G +YK TL A K+L T G E+ I
Sbjct: 806 LNKVLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETI 865
Query: 354 AKLQHRNLVKL---------------------LDEEKH-----KHLDWKLRLSIIKGIAR 387
K++HRNL+KL L + H K LDW R +I G A
Sbjct: 866 GKVRHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAH 925
Query: 388 GLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGY 447
GL YLH D ++HRD+K N+LLD ++ P ISDFG+A+ ++ + V GT GY
Sbjct: 926 GLAYLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGY 985
Query: 448 MAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR 479
MAPE A + S +SDV+S+GV++LE+I K+
Sbjct: 986 MAPENAFTTVKSRESDVYSYGVVLLELITRKK 1017
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 125 bits (313), Expect = 3e-28
Identities = 67/195 (34%), Positives = 107/195 (54%), Gaps = 26/195 (13%)
Query: 311 LGEGGFGPVYKGTLPDGTEIAAKRL-SETSGQGLEEFKNEVIFIAKLQHRNLVKLLDEEK 369
+G+GG G VY+G++P+ ++A KRL +G+ F E+ + +++HR++V+LL
Sbjct: 698 IGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSDHGFTAEIQTLGRIRHRHIVRLLGYVA 757
Query: 370 HK-------------------------HLDWKLRLSIIKGIARGLLYLHEDSPLRVIHRD 404
+K HL W+ R + A+GL YLH D ++HRD
Sbjct: 758 NKDTNLLLYEYMPNGSLGELLHGSKGGHLQWETRHRVAVEAAKGLCYLHHDCSPLILHRD 817
Query: 405 LKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSDV 464
+K++N+LLD + ++DFGLA+ + G+YGY+APEYA KSDV
Sbjct: 818 VKSNNILLDSDFEAHVADFGLAKFLVDGAASECMSSIAGSYGYIAPEYAYTLKVDEKSDV 877
Query: 465 FSFGVLVLEIIYGKR 479
+SFGV++LE+I GK+
Sbjct: 878 YSFGVVLLELIAGKK 892
>IRA1_MOUSE (Q62406) Interleukin-1 receptor-associated kinase 1 (EC
2.7.1.37) (IRAK-1) (IRAK) (Pelle-like protein kinase)
(mPLK)
Length = 710
Score = 124 bits (312), Expect = 4e-28
Identities = 82/224 (36%), Positives = 116/224 (51%), Gaps = 40/224 (17%)
Query: 294 PLTVIQQSTDNFSESFKLGEGGFGPVYKGTLPDGTEIAAKRLSETSG----QGLEEFKNE 349
P I Q T NFSE ++GEGGFG VY+ + + T A KRL E + + F E
Sbjct: 201 PFCEISQGTCNFSEELRIGEGGFGCVYRAVMRN-TTYAVKRLKEEADLEWTMVKQSFLTE 259
Query: 350 VIFIAKLQHRNLVKL---------------------LDEEKHKH------LDWKLRLSII 382
V +++ +H N+V L+++ H L W RL I+
Sbjct: 260 VEQLSRFRHPNIVDFAGYCAESGLYCLVYGFLPNGSLEDQLHLQTQACSPLSWPQRLDIL 319
Query: 383 KGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLAR-----AFEKDQCHT- 436
G AR + +LH+DSP +IH D+K+SNVLLD+ + PK+ DFGLAR + Q T
Sbjct: 320 LGTARAIQFLHQDSP-SLIHGDIKSSNVLLDERLMPKLGDFGLARFSRFAGAKASQSSTV 378
Query: 437 -KTKRVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGKR 479
+T V GT Y+ EY G +V +D FSFGV++LE + G+R
Sbjct: 379 ARTSTVRGTLAYLPEEYIKTGRLAVDTDTFSFGVVILETLAGQR 422
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 124 bits (310), Expect = 7e-28
Identities = 76/219 (34%), Positives = 117/219 (52%), Gaps = 28/219 (12%)
Query: 287 NGDLPTIPLTVIQQSTDNFSESFK--LGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLE 344
NG L T N + +F+ +G+GGFG VY G + +G ++A K LSE S QG +
Sbjct: 554 NGPLKTAKRYFKYSEVVNITNNFERVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYK 612
Query: 345 EFKNEVIFIAKLQHRNLVKLL---DEEKHKHL----------------------DWKLRL 379
EF+ EV + ++ H NL L+ +E H L W+ RL
Sbjct: 613 EFRAEVDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSFILSWEERL 672
Query: 380 SIIKGIARGLLYLHEDSPLRVIHRDLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTK 439
I A+GL YLH ++HRD+K +N+LL++++ K++DFGL+R+F + +
Sbjct: 673 KISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQIST 732
Query: 440 RVIGTYGYMAPEYAMAGLFSVKSDVFSFGVLVLEIIYGK 478
V G+ GY+ PEY + KSDV+S GV++LE+I G+
Sbjct: 733 VVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQ 771
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 120 bits (302), Expect = 6e-27
Identities = 72/196 (36%), Positives = 102/196 (51%), Gaps = 27/196 (13%)
Query: 310 KLGEGGFGPVYKGTLPDGTEIAAKRLSETSGQGLEEFKNEVIFIAKLQHRNLVKL---LD 366
+LG G G VYKG L D +A K+L E QG E F+ E+ I ++ H NLV++
Sbjct: 539 ELGRGESGTVYKGVLEDDRHVAVKKL-ENVRQGKEVFQAELSVIGRINHMNLVRIWGFCS 597
Query: 367 EEKHKHL-----------------------DWKLRLSIIKGIARGLLYLHEDSPLRVIHR 403
E H+ L DW+ R +I G+A+GL YLH + VIH
Sbjct: 598 EGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVAKGLAYLHHECLEWVIHC 657
Query: 404 DLKASNVLLDDEMNPKISDFGLARAFEKDQCHTKTKRVIGTYGYMAPEYAMAGLFSVKSD 463
D+K N+LLD PKI+DFGL + + V GT GY+APE+ + + K D
Sbjct: 658 DVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLGYIAPEWVSSLPITAKVD 717
Query: 464 VFSFGVLVLEIIYGKR 479
V+S+GV++LE++ G R
Sbjct: 718 VYSYGVVLLELLTGTR 733
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 60,748,902
Number of Sequences: 164201
Number of extensions: 2720780
Number of successful extensions: 10057
Number of sequences better than 10.0: 1614
Number of HSP's better than 10.0 without gapping: 1136
Number of HSP's successfully gapped in prelim test: 478
Number of HSP's that attempted gapping in prelim test: 6826
Number of HSP's gapped (non-prelim): 2094
length of query: 493
length of database: 59,974,054
effective HSP length: 114
effective length of query: 379
effective length of database: 41,255,140
effective search space: 15635698060
effective search space used: 15635698060
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC147431.1


