

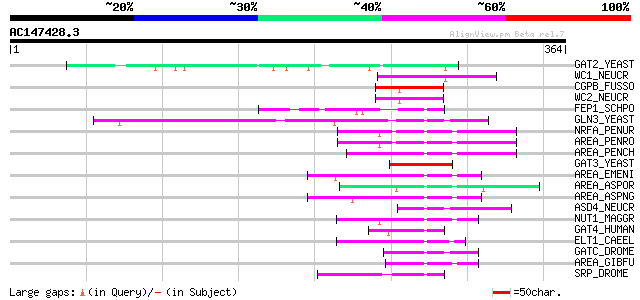
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147428.3 - phase: 0
(364 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
GAT2_YEAST (P40209) GAT2 protein 60 7e-09
WC1_NEUCR (Q01371) White collar 1 protein (WC1) 59 3e-08
CGPB_FUSSO (Q00858) Cutinase gene palindrome-binding protein (PBP) 57 7e-08
WC2_NEUCR (P78714) White collar 2 protein (WC2) 56 2e-07
FEP1_SCHPO (Q10134) Iron-sensing transcription factor 1 (Transcr... 52 3e-06
GLN3_YEAST (P18494) Nitrogen regulatory protein GLN3 50 1e-05
NRFA_PENUR (Q92269) Nitrogen regulatory protein NRFA 49 2e-05
AREA_PENRO (O13508) Nitrogen regulatory protein areA (Nitrogen r... 49 2e-05
AREA_PENCH (Q01582) Nitrogen regulatory protein areA (Nitrogen r... 49 2e-05
GAT3_YEAST (Q07928) GAT3 protein 48 3e-05
AREA_EMENI (P17429) Nitrogen regulatory protein areA 47 6e-05
AREA_ASPOR (O13415) Nitrogen regulatory protein areA 47 6e-05
AREA_ASPNG (O13412) Nitrogen regulatory protein areA 47 1e-04
ASD4_NEUCR (Q9HEV5) GATA type zinc finger protein Asd4 (Ascus de... 46 1e-04
NUT1_MAGGR (Q01168) Nitrogen regulatory protein NUT1 45 2e-04
GAT4_HUMAN (P43694) Transcription factor GATA-4 (GATA binding fa... 45 2e-04
ELT1_CAEEL (P28515) Transcription factor elt-1 45 3e-04
GATC_DROME (P91623) GATA-binding factor-C (Transcription factor ... 45 4e-04
AREA_GIBFU (P78688) Nitrogen regulatory protein areA 44 5e-04
SRP_DROME (P52172) Box A-binding factor (ABF) (Serpent protein) ... 44 8e-04
>GAT2_YEAST (P40209) GAT2 protein
Length = 560
Score = 60.5 bits (145), Expect = 7e-09
Identities = 81/296 (27%), Positives = 112/296 (37%), Gaps = 53/296 (17%)
Query: 38 NNNNNLTQPMEAQEFFQSDNNSNTNNVNTAASDHFIVEDLFDFSNEDVAIEDPTFEE--- 94
+N+ + Q + Q F N+ T A+ L D E ++ E+
Sbjct: 239 SNSTHFRQSIATQNFHSLQNHITTIENRLASL-------LTDRQQEQQQLKQQESEKESS 291
Query: 95 SPPTNSNDSPPLE--TNPTSN----FFTDNSCQNSADGPFSGELSVP-YDDLAELEWVSK 147
SP +N P L+ T+ S F DN S S L P Y A L S
Sbjct: 292 SPFSNKIKLPSLQELTDSISTQHLPTFYDNKRHASDTDLKSSTLHGPLYHRHAFLSTSSS 351
Query: 148 FAEESFSSEDLHKLQLISGLKAPN----NVASKPYEE----SNPTVHSQVSVPAK----- 194
+ S L KLQ + PN N++S P+ N T+ V K
Sbjct: 352 SPSPTAGSAPLQKLQ-VPRQDDPNDKKMNISSSPFNSITYIPNTTLSPMVQTQLKNLTTS 410
Query: 195 ---ARSKRSRVPPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMA----PPKKPSPRKRDPN 247
+ K +R P R + PT TT++ +S+ A P S K +PN
Sbjct: 411 NLNTKKKNNRGRP-----RAIQRQPTLTTSSHFINNSNPGAAAVSTTTPAANSDEK-NPN 464
Query: 248 DGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRY---------KSGRLVPEYR 294
C HC +TP+WR GP G +TLCNACG+ Y KS L+ YR
Sbjct: 465 AKKIIEFCFHCGETETPEWRKGPYGTRTLCNACGLFYRKVTKKFGSKSSNLLLRYR 520
>WC1_NEUCR (Q01371) White collar 1 protein (WC1)
Length = 1167
Score = 58.5 bits (140), Expect = 3e-08
Identities = 30/80 (37%), Positives = 42/80 (52%), Gaps = 2/80 (2%)
Query: 242 RKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRY--KSGRLVPEYRPAASP 299
RKR G R C +C T TP+WR GP G + LCN+CG+R+ ++GR+ P
Sbjct: 921 RKRRKGGGNMVRDCANCHTRNTPEWRRGPSGNRDLCNSCGLRWAKQTGRVSPRTSSRGGN 980
Query: 300 TFVLTKHSNSHRKVQELRRQ 319
++K SNS L R+
Sbjct: 981 GDSMSKKSNSPSHSSPLHRE 1000
>CGPB_FUSSO (Q00858) Cutinase gene palindrome-binding protein (PBP)
Length = 457
Score = 57.0 bits (136), Expect = 7e-08
Identities = 25/52 (48%), Positives = 32/52 (61%), Gaps = 8/52 (15%)
Query: 241 PRKRDPNDGGEGRK--------CLHCATDKTPQWRTGPLGPKTLCNACGVRY 284
P +RDP G + +K C C T +P+WR GP GPKTLCNACG+R+
Sbjct: 380 PLERDPRTGDKKKKLKTSEEYVCTDCGTLDSPEWRKGPSGPKTLCNACGLRW 431
>WC2_NEUCR (P78714) White collar 2 protein (WC2)
Length = 530
Score = 55.8 bits (133), Expect = 2e-07
Identities = 25/52 (48%), Positives = 31/52 (59%), Gaps = 8/52 (15%)
Query: 241 PRKRDPNDGGEGRK--------CLHCATDKTPQWRTGPLGPKTLCNACGVRY 284
P RDP G + +K C C T +P+WR GP GPKTLCNACG+R+
Sbjct: 446 PLDRDPRTGEKKKKIKVAEEYVCTDCGTLDSPEWRKGPSGPKTLCNACGLRW 497
>FEP1_SCHPO (Q10134) Iron-sensing transcription factor 1
(Transcription factor gaf2) (Gaf-2)
Length = 564
Score = 51.6 bits (122), Expect = 3e-06
Identities = 41/127 (32%), Positives = 59/127 (46%), Gaps = 25/127 (19%)
Query: 164 ISGLKAPNNVASKPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRLLVLSPTTTTTTT 223
I L A + + + NP+ SVP+ +K S P S+ ++S TTT T+
Sbjct: 95 IRSLNASKSQSGRKSLSPNPS-----SVPSSTETKASPTP---LESKPQIVSDTTTETSN 146
Query: 224 TTT---SSHS--DTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCN 278
T+ SSH+ + +PP +PS C +CAT TP WR G +CN
Sbjct: 147 GTSRRRSSHNQHEDSSPPHEPSVTF-----------CQNCATTNTPLWRRDESG-NPICN 194
Query: 279 ACGVRYK 285
ACG+ YK
Sbjct: 195 ACGLYYK 201
Score = 33.1 bits (74), Expect = 1.1
Identities = 15/34 (44%), Positives = 18/34 (52%), Gaps = 1/34 (2%)
Query: 252 GRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYK 285
G+ C +C T WR GP LCNACG+ K
Sbjct: 9 GQSCSNCHKTTTSLWRRGP-DNSLLCNACGLYQK 41
>GLN3_YEAST (P18494) Nitrogen regulatory protein GLN3
Length = 730
Score = 49.7 bits (117), Expect = 1e-05
Identities = 58/265 (21%), Positives = 113/265 (41%), Gaps = 20/265 (7%)
Query: 56 DNNSNTNNVNTAASD--HFIVEDLFDFSNEDVAIEDPTFEESPPTNSNDSPPLETNPTSN 113
D+ + T ++ AAS+ + + L+DF+ + + S ++ +S + ++
Sbjct: 111 DSLATTQPIDIAASNQQNGEIAQLWDFNVDQFNMTPSNSSGSATISAPNSFTSDIPQYNH 170
Query: 114 FFTDNSCQNSADGPFSGELSVPYDDLAELEWVSKFAEESFSSEDLHKLQLISGLKAPNNV 173
NS S+ P++ S + + S +S S +++KLQ + + N+
Sbjct: 171 GSLGNSVSKSSLFPYNSSTSNSNINQPSINNNSNTNAQSHHSFNIYKLQNNNSSSSAMNI 230
Query: 174 ASKPYEESNPTVHSQVSVPAKARSKRSRVPPCNWTSRL----LVLSPTTTTTTTTTTSSH 229
+ +N + +S + P +S + N T+ + L+ ++T+ ++
Sbjct: 231 TN-----NNNSNNSNIQHPFLKKSDSIGLSSSNTTNSVRKNSLIKPMSSTSLANFKRAAS 285
Query: 230 SDTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRL 289
+ +PS + + P +C +C T KTP WR P G TLCNACG+ K L
Sbjct: 286 VSSSISNMEPSGQNKKPLI-----QCFNCKTFKTPLWRRSPEG-NTLCNACGLFQK---L 336
Query: 290 VPEYRPAASPTFVLTKHSNSHRKVQ 314
RP + + V+ K + R Q
Sbjct: 337 HGTMRPLSLKSDVIKKRISKKRAKQ 361
>NRFA_PENUR (Q92269) Nitrogen regulatory protein NRFA
Length = 865
Score = 49.3 bits (116), Expect = 2e-05
Identities = 35/120 (29%), Positives = 53/120 (44%), Gaps = 9/120 (7%)
Query: 216 PTTTTTTTTTTSSHSDTMAPPKKPSP---RKRDPNDGGEGRKCLHCATDKTPQWRTGPLG 272
PT + +T S + P + SP + DPN G C +C T TP WR P G
Sbjct: 625 PTNHPSPSTPPESGLSSAVPSRPGSPGGSKNGDPNAGPT--TCTNCFTQTTPLWRRNPEG 682
Query: 273 PKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQ 332
+ LCNACG+ K +V RP + T V+ K + S + + ++ + +Q
Sbjct: 683 -QPLCNACGLFLKLHGVV---RPLSLKTDVIKKRNRSSANTLAVGTSRSSKKSSRKNSIQ 738
>AREA_PENRO (O13508) Nitrogen regulatory protein areA (Nitrogen
regulator nmc)
Length = 860
Score = 49.3 bits (116), Expect = 2e-05
Identities = 35/120 (29%), Positives = 53/120 (44%), Gaps = 9/120 (7%)
Query: 216 PTTTTTTTTTTSSHSDTMAPPKKPSP---RKRDPNDGGEGRKCLHCATDKTPQWRTGPLG 272
PT + +T S + P + SP + DPN G C +C T TP WR P G
Sbjct: 620 PTNHPSPSTPPESGLSSAVPSRPGSPGGSKNGDPNAGPT--TCTNCFTQTTPLWRRNPEG 677
Query: 273 PKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQ 332
+ LCNACG+ K +V RP + T V+ K + S + + ++ + +Q
Sbjct: 678 -QPLCNACGLFLKLHGVV---RPLSLKTDVIKKRNRSSASTLAVGTSRSSKKSSRKNSIQ 733
>AREA_PENCH (Q01582) Nitrogen regulatory protein areA (Nitrogen
regulator nre)
Length = 725
Score = 49.3 bits (116), Expect = 2e-05
Identities = 31/111 (27%), Positives = 48/111 (42%), Gaps = 4/111 (3%)
Query: 222 TTTTTSSHSDTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACG 281
+T S S P ++P K + G C +C T TP WR P G + LCNACG
Sbjct: 492 STLPESGLSSRCLPARQPGGLKNGSTNAGPEPACTNCFTQTTPLWRRNPEG-QPLCNACG 550
Query: 282 VRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQELRRQKEMMRAQQHQLLQ 332
+ K +V RP + T V+ K + S + + ++ + +Q
Sbjct: 551 LFLKLHGVV---RPLSLKTDVIKKRNRSSANTLTVGTSRSSKKSSRKNSIQ 598
>GAT3_YEAST (Q07928) GAT3 protein
Length = 141
Score = 48.1 bits (113), Expect = 3e-05
Identities = 24/42 (57%), Positives = 27/42 (64%), Gaps = 1/42 (2%)
Query: 250 GEGRKCLHCATDKT-PQWRTGPLGPKTLCNACGVRYKSGRLV 290
G R+C CA KT PQWR GP G TLCNACG+ Y+ LV
Sbjct: 67 GVTRRCPQCAVIKTSPQWREGPDGEVTLCNACGLFYRKIFLV 108
>AREA_EMENI (P17429) Nitrogen regulatory protein areA
Length = 876
Score = 47.4 bits (111), Expect = 6e-05
Identities = 35/117 (29%), Positives = 55/117 (46%), Gaps = 7/117 (5%)
Query: 196 RSKRSRVPPCNWTSRLL---VLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEG 252
R K +R T++LL + + ++ T+ T S ++ AP + SP +
Sbjct: 611 RQKIARTSSTPNTAQLLRQSMQNQSSHTSPNTPPESGLNSAAPSRPASPGGTKNGEQNGP 670
Query: 253 RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNS 309
C +C T TP WR P G + LCNACG+ K +V RP + T V+ K + +
Sbjct: 671 TTCTNCFTQTTPLWRRNPEG-QPLCNACGLFLKLHGVV---RPLSLKTDVIKKRNRN 723
>AREA_ASPOR (O13415) Nitrogen regulatory protein areA
Length = 866
Score = 47.4 bits (111), Expect = 6e-05
Identities = 39/142 (27%), Positives = 57/142 (39%), Gaps = 15/142 (10%)
Query: 217 TTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEG--RKCLHCATDKTPQWRTGPLGPK 274
T+ T+ T S + P + SP D G C +C T TP WR P G +
Sbjct: 624 TSHTSPNTPPESALSSAVPSRPASPGGSKNGDQGSNGPTTCTNCFTQTTPLWRRNPEG-Q 682
Query: 275 TLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNS---------HRKVQELRRQKEMMRA 325
LCNACG+ K +V RP + T V+ K + S R ++ R+ + +A
Sbjct: 683 PLCNACGLFLKLHGVV---RPLSLKTDVIKKRNRSSANSLAVGTSRASKKTARKNSVQQA 739
Query: 326 QQHQLLQLQHHHSIMFEGPSNG 347
+ + FE P G
Sbjct: 740 SVTTPTSSRAQNGTSFESPPAG 761
>AREA_ASPNG (O13412) Nitrogen regulatory protein areA
Length = 882
Score = 46.6 bits (109), Expect = 1e-04
Identities = 38/118 (32%), Positives = 52/118 (43%), Gaps = 8/118 (6%)
Query: 196 RSKRSRVPPCNWTSRLLVLSPTTTTTTT---TTTSSHSDTMAPPKKPSPRKRDPNDGGEG 252
R K +R T++LL S T+ T T S + P + SP + G
Sbjct: 613 RQKIARTTSTPNTAQLLRQSMNANTSHTSPNTPPESGLSSAVPSRPASPGGSKNGEQSSG 672
Query: 253 -RKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNS 309
C +C T TP WR P G + LCNACG+ K +V RP + T V+ K + S
Sbjct: 673 PTTCTNCFTQTTPLWRRNPEG-QPLCNACGLFLKLHGVV---RPLSLKTDVIKKRNRS 726
>ASD4_NEUCR (Q9HEV5) GATA type zinc finger protein Asd4 (Ascus
development protein 4)
Length = 426
Score = 46.2 bits (108), Expect = 1e-04
Identities = 27/75 (36%), Positives = 38/75 (50%), Gaps = 4/75 (5%)
Query: 255 CLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHSNSHRKVQ 314
C +CAT TP WR +G + LCNACG+ K L RP + T V+ +
Sbjct: 16 CQNCATSTTPLWRRDEMG-QVLCNACGLFLK---LHGRPRPISLKTDVIKSRNRVKTMRP 71
Query: 315 ELRRQKEMMRAQQHQ 329
+L +QK+ + QQ Q
Sbjct: 72 DLAKQKKQQQQQQQQ 86
>NUT1_MAGGR (Q01168) Nitrogen regulatory protein NUT1
Length = 956
Score = 45.4 bits (106), Expect = 2e-04
Identities = 33/104 (31%), Positives = 46/104 (43%), Gaps = 15/104 (14%)
Query: 215 SPTTTTTTTTTTSSHSDTMAPPKKPSP----------RKRDPNDGGEG-RKCLHCATDKT 263
SP T+ S + P P P +++ N GG+ C +CAT T
Sbjct: 612 SPPADMANGHTSGFSSVVHSRPSSPPPGSKNGSTTNLQQQGNNQGGDAPTTCTNCATQTT 671
Query: 264 PQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKHS 307
P WR P G + LCNACG+ K +V RP + T V+ K +
Sbjct: 672 PLWRRNPEG-QPLCNACGLFLKLHGVV---RPLSLKTDVIKKRN 711
>GAT4_HUMAN (P43694) Transcription factor GATA-4 (GATA binding
factor-4)
Length = 442
Score = 45.4 bits (106), Expect = 2e-04
Identities = 23/55 (41%), Positives = 30/55 (53%), Gaps = 6/55 (10%)
Query: 236 PKKPSPRKRDPN-----DGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYK 285
P + +P R PN D EGR+C++C TP WR G LCNACG+ +K
Sbjct: 193 PGRANPAARHPNLDMFDDFSEGRECVNCGAMSTPLWRRDGTG-HYLCNACGLYHK 246
Score = 32.0 bits (71), Expect = 2.5
Identities = 16/45 (35%), Positives = 22/45 (48%), Gaps = 1/45 (2%)
Query: 241 PRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYK 285
P++R G C +C T T WR G + +CNACG+ K
Sbjct: 257 PQRRLSASRRVGLSCANCQTTTTTLWRRNAEG-EPVCNACGLYMK 300
>ELT1_CAEEL (P28515) Transcription factor elt-1
Length = 416
Score = 45.1 bits (105), Expect = 3e-04
Identities = 24/85 (28%), Positives = 41/85 (48%), Gaps = 4/85 (4%)
Query: 215 SPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEGRKCLHCATDKTPQWRTGPLGPK 274
S + ++++ +TS+ +T++ + S + E R+C++C TP WR G
Sbjct: 177 SSSASSSSANSTSTPKNTISKANRSSGGANNSQFSTEDRECVNCGVHNTPLWRRDGSG-N 235
Query: 275 TLCNACGVRYKSGRLVPEYRPAASP 299
LCNACG+ +K RP P
Sbjct: 236 YLCNACGLYFKMNH---HARPLVKP 257
Score = 35.8 bits (81), Expect = 0.17
Identities = 18/46 (39%), Positives = 26/46 (56%), Gaps = 2/46 (4%)
Query: 241 PRKRDPN-DGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYK 285
P+KR N G +C++C T+ T WR G +CNACG+ +K
Sbjct: 257 PKKRQQNAQKRTGIECVNCRTNTTTLWRRNGEG-HPVCNACGLYFK 301
>GATC_DROME (P91623) GATA-binding factor-C (Transcription factor
GATA-C) (dGATA-C) (Grain protein)
Length = 486
Score = 44.7 bits (104), Expect = 4e-04
Identities = 25/62 (40%), Positives = 31/62 (49%), Gaps = 4/62 (6%)
Query: 246 PNDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTK 305
P EGR+C++C TP WR G LCNACG+ YK + + RP P LT
Sbjct: 252 PKQREEGRECVNCGATSTPLWRRDGTG-HYLCNACGLYYK---MNGQNRPLIKPKRRLTL 307
Query: 306 HS 307
S
Sbjct: 308 QS 309
Score = 35.0 bits (79), Expect = 0.30
Identities = 15/34 (44%), Positives = 19/34 (55%), Gaps = 1/34 (2%)
Query: 252 GRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYK 285
G C +C T T WR G + +CNACG+ YK
Sbjct: 318 GTSCANCKTTTTTLWRRNASG-EPVCNACGLYYK 350
>AREA_GIBFU (P78688) Nitrogen regulatory protein areA
Length = 971
Score = 44.3 bits (103), Expect = 5e-04
Identities = 25/61 (40%), Positives = 32/61 (51%), Gaps = 4/61 (6%)
Query: 247 NDGGEGRKCLHCATDKTPQWRTGPLGPKTLCNACGVRYKSGRLVPEYRPAASPTFVLTKH 306
NDG C +C T TP WR P G + LCNACG+ K +V RP + T V+ K
Sbjct: 686 NDGNAPTTCTNCFTQTTPLWRRNPEG-QPLCNACGLFLKLHGVV---RPLSLKTDVIKKR 741
Query: 307 S 307
+
Sbjct: 742 N 742
>SRP_DROME (P52172) Box A-binding factor (ABF) (Serpent protein)
(GATA-binding factor-B) (Transcription factor GATA-B)
(dGATA-B)
Length = 779
Score = 43.5 bits (101), Expect = 8e-04
Identities = 28/83 (33%), Positives = 36/83 (42%), Gaps = 7/83 (8%)
Query: 203 PPCNWTSRLLVLSPTTTTTTTTTTSSHSDTMAPPKKPSPRKRDPNDGGEGRKCLHCATDK 262
P N TS + T T TT + A ++ S KR G C +C T
Sbjct: 273 PGSNATSAATSAVASGTAATAATTLDEHVSRANSRRLSASKR------AGLSCSNCHTTH 326
Query: 263 TPQWRTGPLGPKTLCNACGVRYK 285
T WR P G + +CNACG+ YK
Sbjct: 327 TSLWRRNPAG-EPVCNACGLYYK 348
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.129 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,347,972
Number of Sequences: 164201
Number of extensions: 2119753
Number of successful extensions: 10037
Number of sequences better than 10.0: 224
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 165
Number of HSP's that attempted gapping in prelim test: 7934
Number of HSP's gapped (non-prelim): 1348
length of query: 364
length of database: 59,974,054
effective HSP length: 112
effective length of query: 252
effective length of database: 41,583,542
effective search space: 10479052584
effective search space used: 10479052584
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC147428.3


