

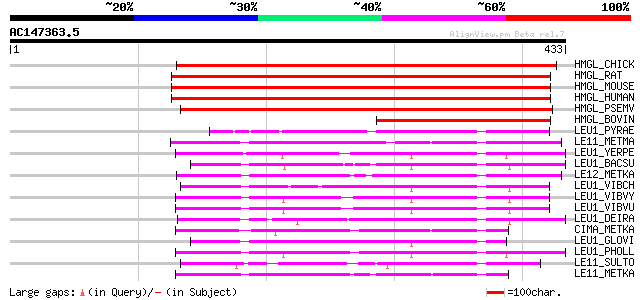
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147363.5 - phase: 0
(433 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HMGL_CHICK (P35915) Hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4)... 398 e-110
HMGL_RAT (P97519) Hydroxymethylglutaryl-CoA lyase, mitochondrial... 395 e-109
HMGL_MOUSE (P38060) Hydroxymethylglutaryl-CoA lyase, mitochondri... 389 e-107
HMGL_HUMAN (P35914) Hydroxymethylglutaryl-CoA lyase, mitochondri... 387 e-107
HMGL_PSEMV (P13703) Hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4)... 309 1e-83
HMGL_BOVIN (Q29448) Hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4)... 175 2e-43
LEU1_PYRAE (Q8ZW35) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 72 4e-12
LE11_METMA (P58967) 2-isopropylmalate synthase 1 (EC 2.3.3.13) (... 69 2e-11
LEU1_YERPE (Q8ZIG8) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 66 2e-10
LEU1_BACSU (P94565) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 66 2e-10
LE12_METKA (Q8TYM1) 2-isopropylmalate synthase 2 (EC 2.3.3.13) (... 66 2e-10
LEU1_VIBCH (Q9KP83) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 66 2e-10
LEU1_VIBVY (Q7MP77) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 65 3e-10
LEU1_VIBVU (Q8DEE1) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 65 3e-10
LEU1_DEIRA (Q9RUA9) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 65 4e-10
CIMA_METKA (Q8TYB1) (R)-citramalate synthase (EC 2.3.3.-) 65 4e-10
LEU1_GLOVI (Q7NI93) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 64 6e-10
LEU1_PHOLL (Q7N129) 2-isopropylmalate synthase (EC 2.3.3.13) (Al... 64 8e-10
LE11_SULTO (Q974X3) 2-isopropylmalate synthase 1 (EC 2.3.3.13) (... 64 1e-09
LE11_METKA (Q8TW28) 2-isopropylmalate synthase 1 (EC 2.3.3.13) (... 64 1e-09
>HMGL_CHICK (P35915) Hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4)
(HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA
lyase)
Length = 298
Score = 398 bits (1023), Expect = e-110
Identities = 199/296 (67%), Positives = 237/296 (79%)
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAK 190
P+ VK+VEVGPRDGLQNEK++VPT VKI LI L+ TGL VIEATSFVSP+WVPQ+AD
Sbjct: 3 PQRVKVVEVGPRDGLQNEKSVVPTPVKIRLIDMLSETGLPVIEATSFVSPRWVPQMADHA 62
Query: 191 DVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSR 250
+VMQ ++ L G+ PVLTPNLKGF+AAVAAGA+EV++F +ASE F+K NINCSIEESL R
Sbjct: 63 EVMQGINKLPGVSYPVLTPNLKGFQAAVAAGAKEVSIFGAASELFTKKNINCSIEESLER 122
Query: 251 YRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVG 310
+ V AA+ SIPVRGYVSCV+GCP EG + +KVA V+K +Y MGC+EISLGD IG+G
Sbjct: 123 FSEVMNAARAASIPVRGYVSCVLGCPYEGNISAAKVAEVSKKMYSMGCYEISLGDRIGIG 182
Query: 311 TPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYA 370
TPG++ ML AVM VP LAVH HDTYGQ+L NILV+LQMG+S VD+SVAGLGGCPYA
Sbjct: 183 TPGSMKEMLAAVMKEVPVGALAVHCHDTYGQALANILVALQMGVSVVDASVAGLGGCPYA 242
Query: 371 KGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNRV 426
+GASGNVATED+VYMLNGLGI T VD+ KLM G FI L R + SK + A R+
Sbjct: 243 QGASGNVATEDLVYMLNGLGIHTGVDLQKLMDTGTFICNALNRRTNSKVSQAACRL 298
>HMGL_RAT (P97519) Hydroxymethylglutaryl-CoA lyase, mitochondrial
precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL)
(3-hydroxy-3-methylglutarate-CoA lyase)
Length = 325
Score = 395 bits (1014), Expect = e-109
Identities = 199/296 (67%), Positives = 234/296 (78%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEK+IVPT VKI+LI L+ GL VIEATSFVSPKWVPQ+
Sbjct: 26 MGTLPKRVKIVEVGPRDGLQNEKSIVPTPVKIKLIDMLSEAGLPVIEATSFVSPKWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD DV++ + GI PVLTPN+KGFE AVAAGA+EV++F +ASE F++ N+NCSIEE
Sbjct: 86 ADHSDVLKGIQKFPGINYPVLTPNMKGFEEAVAAGAKEVSIFGAASELFTRKNVNCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ V +AA+ SI VRGYVSC +GCP EG V P+KVA VAK LY MGC+EISLGDT
Sbjct: 146 SFQRFDGVMQAARAASISVRGYVSCALGCPYEGKVSPAKVAEVAKKLYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AV+ VP LAVH HDTYGQ+L N LV+LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGLMKDMLTAVLHEVPVAALAVHCHDTYGQALANTLVALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYAKGASGN+ATED+VYML GLGI T V++ KL+ AGDFI + L R + SK A A
Sbjct: 266 CPYAKGASGNLATEDLVYMLTGLGIHTGVNLQKLLEAGDFICQALNRKTSSKVAQA 321
>HMGL_MOUSE (P38060) Hydroxymethylglutaryl-CoA lyase, mitochondrial
precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL)
(3-hydroxy-3-methylglutarate-CoA lyase)
Length = 325
Score = 389 bits (998), Expect = e-107
Identities = 198/296 (66%), Positives = 229/296 (76%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEK+IVPT VKI LI L+ GL VIEATSFVSP WVPQ+
Sbjct: 26 MGTLPKQVKIVEVGPRDGLQNEKSIVPTPVKIRLIDMLSEAGLPVIEATSFVSPNWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
AD DV++ + GI PVLTPN+KGFE AVAAGA+EV+VF + SE F++ N NCSIEE
Sbjct: 86 ADHSDVLKGIQKFPGINYPVLTPNMKGFEEAVAAGAKEVSVFGAVSELFTRKNANCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ V +AA+ SI VRGYVSC +GCP EG V P+KVA VAK LY MGC+EISLGDT
Sbjct: 146 SFQRFAGVMQAAQAASISVRGYVSCALGCPYEGKVSPAKVAEVAKKLYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AVM VP L VH HDT GQ+L N LV+LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGLMKDMLTAVMHEVPVTALGVHCHDTIGQALANTLVALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYAKGASGN+ATED+VYMLNGLGI T V++ KL+ AGDFI + L R + SK A A
Sbjct: 266 CPYAKGASGNLATEDLVYMLNGLGIHTGVNLQKLLEAGDFICQALNRKTSSKVAQA 321
>HMGL_HUMAN (P35914) Hydroxymethylglutaryl-CoA lyase, mitochondrial
precursor (EC 4.1.3.4) (HMG-CoA lyase) (HL)
(3-hydroxy-3-methylglutarate-CoA lyase)
Length = 325
Score = 387 bits (995), Expect = e-107
Identities = 195/296 (65%), Positives = 231/296 (77%)
Query: 127 MKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQL 186
M +P+ VKIVEVGPRDGLQNEKNIV T VKI+LI L+ GLSVIE TSFVSPKWVPQ+
Sbjct: 26 MGTLPKRVKIVEVGPRDGLQNEKNIVSTPVKIKLIDMLSEAGLSVIETTSFVSPKWVPQM 85
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
D +V++ + GI PVLTPNLKGFEAAVAAGA+EV +F +ASE F+K NINCSIEE
Sbjct: 86 GDHTEVLKGIQKFPGINYPVLTPNLKGFEAAVAAGAKEVVIFGAASELFTKKNINCSIEE 145
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDT 306
S R+ A+ +AA+ +I VRGYVSC +GCP EG + P+KVA V K Y MGC+EISLGDT
Sbjct: 146 SFQRFDAILKAAQSANISVRGYVSCALGCPYEGKISPAKVAEVTKKFYSMGCYEISLGDT 205
Query: 307 IGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
IGVGTPG + ML AVM VP LAVH HDTYGQ+L N L++LQMG+S VDSSVAGLGG
Sbjct: 206 IGVGTPGIMKDMLSAVMQEVPLAALAVHCHDTYGQALANTLMALQMGVSVVDSSVAGLGG 265
Query: 367 CPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
CPYA+GASGN+ATED+VYML GLGI T V++ KL+ AG+FI + L R + SK A A
Sbjct: 266 CPYAQGASGNLATEDLVYMLEGLGIHTGVNLQKLLEAGNFICQALNRKTSSKVAQA 321
>HMGL_PSEMV (P13703) Hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4)
(HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA
lyase)
Length = 301
Score = 309 bits (791), Expect = 1e-83
Identities = 155/290 (53%), Positives = 200/290 (68%)
Query: 134 VKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVM 193
VK+ EVGPRDGLQNE+ + ++ LI LA TGL IEA +FVSP+WVPQ+A + +V+
Sbjct: 4 VKVFEVGPRDGLQNERQPLSVAARVGLIGELAGTGLRHIEAGAFVSPRWVPQMAGSDEVL 63
Query: 194 QAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRA 253
+ + + G+ L PN +GFEAA AG REVAVFA+ASE+FS++NINCSI+ES R+
Sbjct: 64 RQLPSNDGVSYTALVPNRQGFEAAQRAGCREVAVFAAASEAFSRNNINCSIDESFERFTP 123
Query: 254 VTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVGTPG 313
V RAA E SI VRGYVSCV+GCP G V P VA VA+ LY++GC+EISLGDTIG G P
Sbjct: 124 VLRAANEASIRVRGYVSCVLGCPFSGAVAPEAVAKVARRLYELGCYEISLGDTIGAGRPD 183
Query: 314 TVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGA 373
+ +P LA HFHDT+G ++ N+ +L G+ DSSVAGLGGCPY+ GA
Sbjct: 184 ETAQLFELCARQLPVAALAGHFHDTWGMAIANVHAALAQGVRTFDSSVAGLGGCPYSPGA 243
Query: 374 SGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIAL 423
SGNVATED++Y+L+GLG T VD+ + G I QLG + S+ +AL
Sbjct: 244 SGNVATEDLLYLLHGLGYSTGVDLEAVAQVGVRISAQLGTANRSRAGLAL 293
>HMGL_BOVIN (Q29448) Hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4)
(HMG-CoA lyase) (HL) (3-hydroxy-3-methylglutarate-CoA
lyase) (Fragment)
Length = 140
Score = 175 bits (444), Expect = 2e-43
Identities = 89/136 (65%), Positives = 104/136 (76%)
Query: 287 AYVAKALYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNI 346
A V K LY MGC+EISLGDTIGVGTPG + ML A + VP LAVH HDTYGQ+L N
Sbjct: 1 AEVTKKLYSMGCYEISLGDTIGVGTPGAMKDMLSAALQEVPVTALAVHCHDTYGQALANT 60
Query: 347 LVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDF 406
L +LQMG+S +DSSVAGLGGCPYA+GASGN+ATED+VYML GLGI T V++ KL+ AG F
Sbjct: 61 LTALQMGVSVMDSSVAGLGGCPYAQGASGNLATEDLVYMLAGLGIHTGVNLQKLLEAGAF 120
Query: 407 IGKQLGRPSGSKTAIA 422
I + L R + SK A A
Sbjct: 121 ICQALNRRTNSKVAQA 136
>LEU1_PYRAE (Q8ZW35) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 364
Score = 71.6 bits (174), Expect = 4e-12
Identities = 65/266 (24%), Positives = 116/266 (43%), Gaps = 17/266 (6%)
Query: 157 KIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLRGIRLPVLTPNLKGFEA 216
K+E+ L G+ +IEA F + + A K + + V + + L +T + +A
Sbjct: 11 KLEIAMALDDAGVDMIEA-GFAAVSDDEKEA-IKLISKEVSRAKVVSLARMTKS--DVDA 66
Query: 217 AVAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTRAAKELSIPVRGYVSCVVGCP 276
A A + +F + S+ K + + EE+L R V AK + +
Sbjct: 67 AAEADVDMIHLFIATSDIHLKYKLGITREEALRRIEEVVSYAKSYGVEILFSAEDATRSD 126
Query: 277 VEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFH 336
+E +A K + G EI++ DT+GV TP + ++ + +P + VH H
Sbjct: 127 LEF------LAKAYKTAIEAGADEINVPDTVGVMTPSRMAYLIKYLRERLPPIPMHVHCH 180
Query: 337 DTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASGNVATEDVVYMLNG-LGIKTNV 395
D +G ++ N + +++ G V G +GN A E+VV ++ LG++TN+
Sbjct: 181 DDFGMAVANTVTAIENGADVAQVVVNNFG------ERAGNAALEEVVAAVHYLLGLRTNI 234
Query: 396 DIGKLMSAGDFIGKQLGRPSGSKTAI 421
+ KL S + K G P A+
Sbjct: 235 KLEKLYSLSQLVSKLFGVPVPPNKAV 260
>LE11_METMA (P58967) 2-isopropylmalate synthase 1 (EC 2.3.3.13)
(Alpha-isopropylmalate synthase 1) (Alpha-IPM synthetase
1)
Length = 405
Score = 69.3 bits (168), Expect = 2e-11
Identities = 77/309 (24%), Positives = 132/309 (41%), Gaps = 23/309 (7%)
Query: 126 FMKGMPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQ 185
F+ P ++I +V RDG Q + + K+ + L S G+ VIEA V
Sbjct: 15 FIDYRPLDIEICDVTLRDGEQTPGVVFSKEQKLAVASELDSMGIEVIEAGFPVVS----- 69
Query: 186 LADAKDVMQAVHNLR-GIRLPVLTPNLKG-FEAAVAAGAREVAVFASASESFSKSNINCS 243
AD K++++ + N R+ L+ +KG +AA+ V++F + S+ K + S
Sbjct: 70 -ADEKEIVKEIANQGFNSRICCLSRAVKGDVDAALECDVDIVSIFIAMSDMHLKYKYHRS 128
Query: 244 IEESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSK-VAYVAKALYDMGCFEIS 302
+E+ L + A + + VR PVE K V K Y +S
Sbjct: 129 LEDMLGCAKEAIEYATDHGLKVRFAAEDASRTPVERLKQAFKEVENEYKVQY------VS 182
Query: 303 LGDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVA 362
L DT+G+ P T ++ + V T + +H HD G + N L + + G + ++V
Sbjct: 183 LADTVGILNPTTTNYLVSEIFKSVNT-AICIHCHDDLGMATANTLAAAEAGAKQLHTTVN 241
Query: 363 GLGGCPYAKGASGNVATEDVVYMLNGLGIKTNVDIGKLMSAGDFIGKQLG-RPSGSKTAI 421
+G +GN + E+V+ L D KL S + + + G PS +K +
Sbjct: 242 AIG------ERAGNASLEEVLVALRVQYGIERYDTTKLNSLSEMVSEYSGITPSVNKAVV 295
Query: 422 ALNRVTADA 430
N T ++
Sbjct: 296 GKNAFTHES 304
>LEU1_YERPE (Q8ZIG8) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 520
Score = 66.2 bits (160), Expect = 2e-10
Identities = 76/315 (24%), Positives = 127/315 (40%), Gaps = 27/315 (8%)
Query: 130 MPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADA 189
M + V I + RDG Q + + K+++ L G+ ++E VS
Sbjct: 1 MSQQVIIFDTTLRDGEQALQASLSVKEKLQIALALERMGVDIMEVGFPVSSPG--DFESV 58
Query: 190 KDVMQAVHNLRGIRLPVLTPNL--KGFEAAVAAGAREVAVFASASESFSKSNINCSIEES 247
+ + Q V N R L EA A A + VF + S +S + S ++
Sbjct: 59 RTIAQQVKNSRVCALARCVDKDIDVAAEALRIAEAFRIHVFLATSTLHIESKLKRSFDDV 118
Query: 248 LSR-YRAVTRAAKELSIPVRGYVSCV-VGCPVEGPVPPSKVAYVAKALYDMGCFEISLGD 305
L+ +V RA R Y V C G P + V +A G I++ D
Sbjct: 119 LAMAVHSVKRA--------RNYTDDVEFSCEDAGRTPIDNLCRVVEAAITAGATTINIPD 170
Query: 306 TIGVGTP---GTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVA 362
T+G TP G ++ L + + ++VH HD G S+ N + ++Q G V+ ++
Sbjct: 171 TVGYTTPYQFGGIITDLYERVPNIDKAIISVHCHDDLGMSVANSITAVQAGARQVEGTIN 230
Query: 363 GLGGCPYAKGASGNVATEDVVYML----NGLGIKTNVDIGKLMSAGDFIGKQLGRPSGSK 418
GLG +GN + E+V+ + LG+ TN++ ++ + K P
Sbjct: 231 GLG------ERAGNCSLEEVIMAIKVRHEMLGVHTNINHQEIYRTSQLVSKICNMPIPGN 284
Query: 419 TAIALNRVTADASKI 433
AI + A +S I
Sbjct: 285 KAIVGSNAFAHSSGI 299
>LEU1_BACSU (P94565) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 518
Score = 66.2 bits (160), Expect = 2e-10
Identities = 68/304 (22%), Positives = 125/304 (40%), Gaps = 29/304 (9%)
Query: 142 RDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVMQAVHNLRG 201
RDG Q+ + T K+ + +L G +IEA S + D V + ++
Sbjct: 12 RDGEQSPGVNLNTQEKLAIAKQLERLGADIIEAGFPASSR-----GDFLAVQEIARTIKN 66
Query: 202 IRLPVLTPNLKG-----FEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRAVTR 256
+ L +KG +EA + VF + S+ K + + E+ + R + +
Sbjct: 67 CSVTGLARCVKGDIDAAWEALKDGAQPRIHVFIATSDIHLKHKLKMTREQVIERAVGMVK 126
Query: 257 AAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVGTP---G 313
AKE P+ + S C E P +A + + + D G I+L DT+G P G
Sbjct: 127 YAKE-RFPIVQW-SAEDACRTELPF----LAEIVEKVIDAGASVINLPDTVGYLAPAEYG 180
Query: 314 TVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGA 373
+ + + + KL+ H HD G ++ N L +++ G ++ +V G+G
Sbjct: 181 NIFKYMKENVPNIHKAKLSAHCHDDLGMAVANSLAAIENGADQIECAVNGIG------ER 234
Query: 374 SGNVATEDVVYMLNG----LGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIALNRVTAD 429
+GN A E++ L+ ++T + + ++ D + K G A+ + A
Sbjct: 235 AGNAALEEIAVALHTRKDFYQVETGITLNEIKRTSDLVSKLTGMAVPRNKAVVGDNAFAH 294
Query: 430 ASKI 433
S I
Sbjct: 295 ESGI 298
>LE12_METKA (Q8TYM1) 2-isopropylmalate synthase 2 (EC 2.3.3.13)
(Alpha-isopropylmalate synthase 2) (Alpha-IPM synthetase
2)
Length = 509
Score = 66.2 bits (160), Expect = 2e-10
Identities = 73/304 (24%), Positives = 126/304 (41%), Gaps = 21/304 (6%)
Query: 131 PEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAK 190
P+ V+I + RDG Q + + K+ + +L G+ IEA + + + K
Sbjct: 11 PDEVRIFDTTLRDGEQTPGVALTPEEKLRIARKLDEIGVDTIEAGFAAASE-----GELK 65
Query: 191 DVMQAVHNLRGIRLPVLTPNLKG-FEAAVAAGAREVAVFASASESFSKSNINCSIEESLS 249
+ + + + +KG +AAV A A V + SE K + EE L
Sbjct: 66 AIRRIAREELDAEVCSMARMVKGDVDAAVEAEADAVHIVVPTSEVHVKKKLRMDREEVLE 125
Query: 250 RYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGV 309
R R V A++ + V +S G E + V A + G + DT+GV
Sbjct: 126 RAREVVEYARDHGLTVE--ISTEDGTRTE----LEYLYEVFDACLEAGAERLGYNDTVGV 179
Query: 310 GTPGTVVPMLLAVMAIVPTEK-LAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCP 368
P + + + V + L+VH HD +G + N + +++ G V +V G+G
Sbjct: 180 MAPEGMFLAVKKLRERVGEDVILSVHCHDDFGMATANTVAAVRAGARQVHVTVNGIG--- 236
Query: 369 YAKGASGNVATEDVVYMLNGL-GIKTNVDIGKLMSAGDFIGKQLG-RPSGSKTAIALNRV 426
+GN A E+VV +L L G+ T + +L + + G R +K + N
Sbjct: 237 ---ERAGNAALEEVVVVLEELYGVDTGIRTERLTELSKLVERLTGVRVPPNKAVVGENAF 293
Query: 427 TADA 430
T ++
Sbjct: 294 THES 297
>LEU1_VIBCH (Q9KP83) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 516
Score = 65.9 bits (159), Expect = 2e-10
Identities = 72/298 (24%), Positives = 125/298 (41%), Gaps = 25/298 (8%)
Query: 134 VKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADAKDVM 193
V I + RDG Q + K+++ + L G+ +IEA VS D + V
Sbjct: 5 VIIFDTTLRDGEQALSASLTVKEKLQIAYALERLGVDIIEAGFPVSSP-----GDFESVQ 59
Query: 194 QAVHNLRGIRLPVLTPNL-KGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSR-Y 251
N++ R+ L+ + K +AA A A +VA S S I+ +++ L R Y
Sbjct: 60 TIAKNIKNSRVCALSRAVAKDIDAA--AEALKVAEAFRIHTFISTSTIH--VQDKLRRSY 115
Query: 252 RAVTRAAKELSIPVRGYVSCV-VGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIGVG 310
V A + R Y V C G P + + +A + G I++ DT+G
Sbjct: 116 DDVVAMAVKAVKHARQYTDDVEFSCEDAGRTPIDNLCRMVEAAINAGARTINIPDTVGYT 175
Query: 311 TP---GTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGC 367
P G ++ L + + ++VH HD G S+ N + ++Q G V+ ++ G+G
Sbjct: 176 VPSEFGGIIQTLFNRVPNIDKAIISVHCHDDLGMSVANSIAAIQAGARQVEGTINGIG-- 233
Query: 368 PYAKGASGNVATEDVVYMLNG----LGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAI 421
+GN A E++ ++ LG+ T + ++ + + P S AI
Sbjct: 234 ----ERAGNCALEEIAMIIKTRQELLGVTTGIKHDEISRTSKLVSQLCNMPIQSNKAI 287
>LEU1_VIBVY (Q7MP77) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 515
Score = 65.1 bits (157), Expect = 3e-10
Identities = 69/306 (22%), Positives = 126/306 (40%), Gaps = 33/306 (10%)
Query: 130 MPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADA 189
M + V I + RDG Q + K+++ + L G+ VIEA VS D
Sbjct: 1 MNDQVIIFDTTLRDGEQALSASLTVKEKLQIAYALERLGVDVIEAGFPVSSP-----GDF 55
Query: 190 KDVMQAVHNLRGIRLPVLTPNLK-----GFEAAVAAGAREVAVFASASESFSKSNINCSI 244
+ V N++ R+ L+ ++ EA A A + F S S + + S
Sbjct: 56 ESVQTIARNIKNSRVCALSRAVEKDIDAAAEALKVAEAFRIHTFISTSTIHVQDKLRRSY 115
Query: 245 EESLSR-YRAVTRAAKELSIPVRGYVSCV-VGCPVEGPVPPSKVAYVAKALYDMGCFEIS 302
++ + RAV A K Y V C G P + + +A + G I+
Sbjct: 116 DDVVEMGVRAVKHARK--------YTDDVEFSCEDAGRTPIDNLCRMVEAAINAGARTIN 167
Query: 303 LGDTIGVGTP---GTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDS 359
+ DT+G P G ++ L + + ++VH HD G S+ N + ++Q G V+
Sbjct: 168 IPDTVGYTVPSEFGGIIQTLFNRVPNIDKAIISVHCHDDLGMSVANSIAAVQAGARQVEG 227
Query: 360 SVAGLGGCPYAKGASGNVATEDVVYMLNG----LGIKTNVDIGKLMSAGDFIGKQLGRPS 415
++ G+G +GN + E++ ++ LG++T ++ ++ + + P
Sbjct: 228 TINGIG------ERAGNCSLEEIAMIIKTRQELLGVRTGINHEEIHRTSKLVSQLCNMPI 281
Query: 416 GSKTAI 421
+ AI
Sbjct: 282 QANKAI 287
>LEU1_VIBVU (Q8DEE1) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 515
Score = 65.1 bits (157), Expect = 3e-10
Identities = 69/306 (22%), Positives = 126/306 (40%), Gaps = 33/306 (10%)
Query: 130 MPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADA 189
M + V I + RDG Q + K+++ + L G+ VIEA VS D
Sbjct: 1 MNDQVIIFDTTLRDGEQALSASLTVKEKLQIAYALERLGVDVIEAGFPVSSP-----GDF 55
Query: 190 KDVMQAVHNLRGIRLPVLTPNLK-----GFEAAVAAGAREVAVFASASESFSKSNINCSI 244
+ V N++ R+ L+ ++ EA A A + F S S + + S
Sbjct: 56 ESVQTIARNIKNSRVCALSRAVEKDIDAAAEALKVAEAFRIHTFISTSTIHVQDKLRRSY 115
Query: 245 EESLSR-YRAVTRAAKELSIPVRGYVSCV-VGCPVEGPVPPSKVAYVAKALYDMGCFEIS 302
++ + RAV A K Y V C G P + + +A + G I+
Sbjct: 116 DDVVEMGVRAVKHARK--------YTDDVEFSCEDAGRTPIDNLCRMVEAAINAGARTIN 167
Query: 303 LGDTIGVGTP---GTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDS 359
+ DT+G P G ++ L + + ++VH HD G S+ N + ++Q G V+
Sbjct: 168 IPDTVGYTVPSEFGGIIQTLFNRVPNIDKAIISVHCHDDLGMSVANSIAAVQAGARQVEG 227
Query: 360 SVAGLGGCPYAKGASGNVATEDVVYMLNG----LGIKTNVDIGKLMSAGDFIGKQLGRPS 415
++ G+G +GN + E++ ++ LG++T ++ ++ + + P
Sbjct: 228 TINGIG------ERAGNCSLEEIAMIIKTRQELLGVRTGINHEEIHRTSKLVSQLCNMPI 281
Query: 416 GSKTAI 421
+ AI
Sbjct: 282 QANKAI 287
>LEU1_DEIRA (Q9RUA9) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 522
Score = 64.7 bits (156), Expect = 4e-10
Identities = 73/311 (23%), Positives = 132/311 (41%), Gaps = 25/311 (8%)
Query: 132 EFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEA-TSFVSPKWVPQLADAK 190
+ ++I + RDG Q+ + K+E+ H LA G+ VIEA SP D +
Sbjct: 14 QHIRIFDTTLRDGEQSPGVALNHAQKLEIAHTLARMGVDVIEAGFPIASP------GDLE 67
Query: 191 DVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAR--EVAVFASASESFSKSNINCSIEESL 248
V + +RG PV+T + A + A A+ E A + + S I+ + +L
Sbjct: 68 GVSRIAREVRG---PVITALARANRADIEAAAKGVEAAEKSRIHTFIATSPIHMQKKLNL 124
Query: 249 SRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPP-SKVAYVAKALYDMGCFEISLGDTI 307
+ RA + + + R YV V + ++ + KA + G I++ DT+
Sbjct: 125 EPDAVIERAVEAVRL-ARQYVDDVEFSAEDATRSEWDFLSRIFKAAVEAGATTINVPDTV 183
Query: 308 GVGTPGTVVPMLLAVMAIVPTEK-LAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGG 366
G TP + + + A +P L+ H HD G ++ N + + + G ++ ++ G+G
Sbjct: 184 GYTTPEEMRKLFAYLKAKLPAHVILSSHCHDDLGMAVANSIAAAEGGARQIECTINGIG- 242
Query: 367 CPYAKGASGNVATEDVVYMLNG----LGIKTNVDIGKLMSAGDFIGKQLGRPSGSKTAIA 422
+GN + E++V + G +T + +L A + + G P AI
Sbjct: 243 -----ERAGNASLEEIVMAFHTRRDVYGYETGIRTRELYRASRLVSRLSGMPVQPNKAIV 297
Query: 423 LNRVTADASKI 433
+ A S I
Sbjct: 298 GDNAFAHESGI 308
>CIMA_METKA (Q8TYB1) (R)-citramalate synthase (EC 2.3.3.-)
Length = 499
Score = 64.7 bits (156), Expect = 4e-10
Identities = 67/265 (25%), Positives = 113/265 (42%), Gaps = 27/265 (10%)
Query: 130 MPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADA 189
MP+ V+I + RDG Q + + K+E+ +L G+ IEA V+ +
Sbjct: 1 MPDRVRIFDTTLRDGEQTPGVSLTVEEKVEIARKLDEFGVDTIEAGFPVA---------S 51
Query: 190 KDVMQAVHNLRGIRLPV----LTPNLKG-FEAAVAAGAREVAVFASASESFSKSNINCSI 244
+ +AV + G L L +KG +AA+ A V VF + S+ + + S
Sbjct: 52 EGEFEAVRAIAGEELDAEICGLARCVKGDIDAAIDADVDCVHVFIATSDIHLRYKLEMSR 111
Query: 245 EESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLG 304
EE+L R A + + V + V KA + G +++
Sbjct: 112 EEALERAIEGVEYASDHGVTVE------FSAEDATRTDRDYLLEVYKATVEAGADRVNVP 165
Query: 305 DTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGL 364
DT+GV TP + + V+ V ++VH H+ +G ++ N L +++ G V +V G+
Sbjct: 166 DTVGVMTPPEMYRLTAEVVDAVDV-PVSVHCHNDFGMAVANSLAAVEAGAEQVHVTVNGI 224
Query: 365 GGCPYAKGASGNVATEDVVYMLNGL 389
G +GN + E VV L L
Sbjct: 225 G------ERAGNASLEQVVMALKAL 243
>LEU1_GLOVI (Q7NI93) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 538
Score = 64.3 bits (155), Expect = 6e-10
Identities = 61/252 (24%), Positives = 113/252 (44%), Gaps = 18/252 (7%)
Query: 142 RDGLQNEKNIVPTDVKIELIHRLASTGLSVIEA-TSFVSPKWVPQLADAKDVMQAVHNLR 200
RDG Q+ + D K+E+ +LA G+ VIEA ++ SP D + V + +
Sbjct: 14 RDGEQSPGATLNADEKVEIARQLARLGVDVIEAGFAYASP------GDFEAVERVARTVG 67
Query: 201 GIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEESLSRYRA-VTRAAK 259
PV+ + + + A A + A S + +E L + RA V A
Sbjct: 68 TEDGPVICSLARAIRSDIQAAAEAIRPAARGRIHTFISTSDIHLEHQLRKSRAEVLAIAA 127
Query: 260 ELSIPVRGYVSCVVGCPVE-GPVPPSKVAYVAKALYDMGCFEISLGDTIGVGTP---GTV 315
E+ +G+V V P++ G P + V +A G +++ DT+G TP G +
Sbjct: 128 EMVAFAKGFVDDVEFSPMDAGRSAPEYLYRVLEAAIAAGATTVNIPDTVGYLTPAEFGGL 187
Query: 316 VPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCPYAKGASG 375
+ + + + ++VH H+ G ++ N L +++ G ++ +V G+G +G
Sbjct: 188 IRGITQNVRGIERAVISVHCHNDLGLAVANSLAAIENGARQIECTVNGIG------ERAG 241
Query: 376 NVATEDVVYMLN 387
N + E++V L+
Sbjct: 242 NCSLEEIVMALH 253
>LEU1_PHOLL (Q7N129) 2-isopropylmalate synthase (EC 2.3.3.13)
(Alpha-isopropylmalate synthase) (Alpha-IPM synthetase)
Length = 520
Score = 63.9 bits (154), Expect = 8e-10
Identities = 65/316 (20%), Positives = 129/316 (40%), Gaps = 29/316 (9%)
Query: 130 MPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADA 189
M + V I + RDG Q + + + K+++ L G+ ++E VS D
Sbjct: 1 MNQQVIIFDTTLRDGEQALQASLCVNEKLQIALALERMGVDIMEVGFPVSSP-----GDF 55
Query: 190 KDVMQAVHNLRGIRLPVLTPNLK-----GFEAAVAAGAREVAVFASASESFSKSNINCSI 244
+ V ++ R+ L ++ EA A A + VF + S+ +S + +
Sbjct: 56 ESVQTIAKQIKNSRVCALARCVEKDIDVAAEALKVADAFRIHVFLATSDLHIESKLKKTF 115
Query: 245 EESLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLG 304
E + + A+ + V C G P + + + G I++
Sbjct: 116 ENVMEMATQSIKRARRYTDDVE------FSCEDAGRTPIDNLCRIVENAIKAGATTINIP 169
Query: 305 DTIGVGTP---GTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSV 361
DT+G TP G ++ L+ + + ++VH HD G S+ N + ++Q G ++ ++
Sbjct: 170 DTVGYTTPYQFGGIITNLIERVPNIDKAIISVHCHDDLGMSVANSITAVQSGARQIEGTI 229
Query: 362 AGLGGCPYAKGASGNVATEDVVYML----NGLGIKTNVDIGKLMSAGDFIGKQLGRPSGS 417
GLG +GN + E+V+ + LG+ TN++ ++ + + P +
Sbjct: 230 NGLG------ERAGNCSLEEVIMAIKVRQQMLGVYTNINHQEIYRTSQLVSQLCNMPIPA 283
Query: 418 KTAIALNRVTADASKI 433
AI + + +S I
Sbjct: 284 NKAIVGSNAFSHSSGI 299
>LE11_SULTO (Q974X3) 2-isopropylmalate synthase 1 (EC 2.3.3.13)
(Alpha-isopropylmalate synthase 1) (Alpha-IPM synthetase
1)
Length = 386
Score = 63.5 bits (153), Expect = 1e-09
Identities = 66/292 (22%), Positives = 124/292 (41%), Gaps = 36/292 (12%)
Query: 134 VKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEAT-------SFVSPKWVPQL 186
V+I + RDG Q + + K+++ +LA G+ VIEA F++ K + L
Sbjct: 12 VRIFDTTLRDGEQAPGIDLTVEQKVKIARKLAELGVDVIEAGFPAASEGEFIATKKI--L 69
Query: 187 ADAKDVMQAVHNLRGIRLPVLTPNLKGFEAAVAAGAREVAVFASASESFSKSNINCSIEE 246
+ D ++ + R N + + + G + VF + S+ K + + E+
Sbjct: 70 EEVGDQVEVIGLSRA--------NKQDIDKTIDTGISSIHVFIATSDIHLKYKLKMTREQ 121
Query: 247 SLSRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKAL---YDMGCFEISL 303
L + R AK+ + +V E K ++ KA+ + G I++
Sbjct: 122 VLDKIYESVRYAKDHGL--------IVEYSPEDATRTDK-DFLLKAVSTAIEAGADRINI 172
Query: 304 GDTIGVGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAG 363
DT+GV P ++ V+++ + ++VH H+ +G + N + + G V +V G
Sbjct: 173 PDTVGVMHPFKFHDLIKDVVSVTKDKIVSVHCHNDFGLATANSIAGVMAGARQVHVTVNG 232
Query: 364 LGGCPYAKGASGNVATEDVVYMLNG-LGIKTNVDIGKLMSAGDFIGKQLGRP 414
+G +GN + E+VV L LG + N+ KL + + G P
Sbjct: 233 IG------ERAGNASLEEVVMALKKLLGYEVNIKTYKLYETSRLVSELTGVP 278
>LE11_METKA (Q8TW28) 2-isopropylmalate synthase 1 (EC 2.3.3.13)
(Alpha-isopropylmalate synthase 1) (Alpha-IPM synthetase
1)
Length = 397
Score = 63.5 bits (153), Expect = 1e-09
Identities = 65/261 (24%), Positives = 107/261 (40%), Gaps = 19/261 (7%)
Query: 130 MPEFVKIVEVGPRDGLQNEKNIVPTDVKIELIHRLASTGLSVIEATSFVSPKWVPQLADA 189
+P+ V + + RDG Q + K+E+ H L G+ IEA V + +
Sbjct: 15 LPDEVIVYDTTLRDGEQTPGVSFTPEQKLEIAHLLDELGVQQIEAGFPVVSE-----GER 69
Query: 190 KDVMQAVHNLRGIRLPVLTPNLKG-FEAAVAAGAREVAVFASASESFSKSNINCSIEESL 248
V + H + L L+G +AA+ V F + SE K + S EE L
Sbjct: 70 DAVRRIAHEGLNADILCLARTLRGDVDAALDCDVDGVITFIATSELHLKHKLRMSREEVL 129
Query: 249 SRYRAVTRAAKELSIPVRGYVSCVVGCPVEGPVPPSKVAYVAKALYDMGCFEISLGDTIG 308
R AK+ + V S G E + V + + G + DT+G
Sbjct: 130 ERIADTVEYAKDHGLWVA--FSAEDGTRTEFEF----LERVYRTAEECGADRVHATDTVG 183
Query: 309 VGTPGTVVPMLLAVMAIVPTEKLAVHFHDTYGQSLPNILVSLQMGISAVDSSVAGLGGCP 368
V P + + + +V + VH HD +G ++ N L +++ G A+ ++V G+G
Sbjct: 184 VMIPAAMRLFVAKIREVVDLP-IGVHCHDDFGMAVANSLAAVEAGAQAISTTVNGIG--- 239
Query: 369 YAKGASGNVATEDVVYMLNGL 389
+GN A E+V+ L L
Sbjct: 240 ---ERAGNAALEEVIMALKEL 257
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,533,301
Number of Sequences: 164201
Number of extensions: 2040192
Number of successful extensions: 6159
Number of sequences better than 10.0: 154
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 5968
Number of HSP's gapped (non-prelim): 174
length of query: 433
length of database: 59,974,054
effective HSP length: 113
effective length of query: 320
effective length of database: 41,419,341
effective search space: 13254189120
effective search space used: 13254189120
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147363.5


