

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147179.10 - phase: 1 /pseudo
(254 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
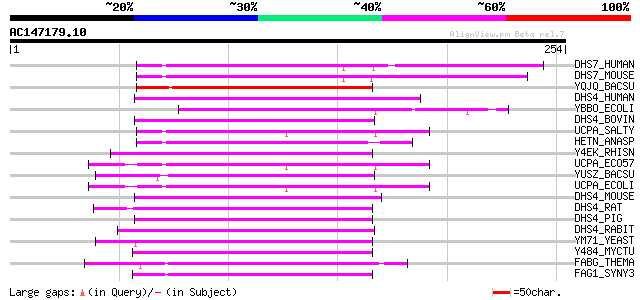
Score E
Sequences producing significant alignments: (bits) Value
DHS7_HUMAN (Q9Y394) Dehydrogenase/reductase SDR family member 7 ... 125 1e-28
DHS7_MOUSE (Q9CXR1) Dehydrogenase/reductase SDR family member 7 ... 112 9e-25
YQJQ_BACSU (P54554) Hypothetical oxidoreductase yqjQ (EC 1.-.-.-) 96 6e-20
DHS4_HUMAN (Q9BTZ2) Dehydrogenase/reductase SDR family member 4 ... 82 1e-15
YBBO_ECOLI (P77388) Hypothetical oxidoreductase ybbO (EC 1.-.-.-) 77 5e-14
DHS4_BOVIN (Q8SPU8) Dehydrogenase/reductase SDR family member 4 ... 77 5e-14
UCPA_SALTY (P37441) Oxidoreductase ucpA (EC 1.-.-.-) 73 8e-13
HETN_ANASP (P37694) Ketoacyl reductase hetN (EC 1.3.1.-) 72 1e-12
Y4EK_RHISN (P55434) Putative short-chain type dehydrogenase/redu... 72 1e-12
UCPA_ECO57 (Q8XBJ4) Oxidoreductase ucpA (EC 1.-.-.-) 72 2e-12
YUSZ_BACSU (P37959) Hypothetical oxidoreductase yusZ (EC 1.-.-.-... 71 3e-12
UCPA_ECOLI (P37440) Oxidoreductase ucpA (EC 1.-.-.-) 71 3e-12
DHS4_MOUSE (Q99LB2) Dehydrogenase/reductase SDR family member 4 ... 71 3e-12
DHS4_RAT (Q8VID1) Dehydrogenase/reductase SDR family member 4 (E... 70 4e-12
DHS4_PIG (Q8WNV7) Dehydrogenase/reductase SDR family member 4 (E... 70 5e-12
DHS4_RABIT (Q9GKX2) Dehydrogenase/reductase SDR family member 4 ... 70 6e-12
YM71_YEAST (Q05016) Hypothetical oxidoreductase in MRPL44-MTF1 i... 69 1e-11
Y484_MYCTU (Q11150) Putative oxidoreductase Rv0484c/MT0502 (EC 1... 67 3e-11
FABG_THEMA (Q9X248) 3-oxoacyl-[acyl-carrier-protein] reductase (... 67 4e-11
FAG1_SYNY3 (P73574) 3-oxoacyl-[acyl-carrier-protein] reductase 1... 66 9e-11
>DHS7_HUMAN (Q9Y394) Dehydrogenase/reductase SDR family member 7
precursor (EC 1.1.-.-) (Retinal short-chain
dehydrogenase/reductase 4) (retSDR4) (CGI-86)
(UNQ285/PRO3448)
Length = 339
Score = 125 bits (314), Expect = 1e-28
Identities = 70/201 (34%), Positives = 112/201 (54%), Gaps = 19/201 (9%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S +D + + + ++N GT++LT+ + P M+ R +G V ++S
Sbjct: 132 IDILVNNGGMSQ-RSLCMDTSLDVYRKLIELNYLGTVSLTKCVLPHMIERKQGKIVTVNS 190
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPIE------------ 165
G P Y ASK+AL G+F+ LR+EL GI V+ +CPGP++
Sbjct: 191 ILGIISVPLSIGYCASKHALRGFFNGLRTELATYPGIIVSNICPGPVQSNIVENSLAGEV 250
Query: 166 --TANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYW 223
T N+G Q +++ +CV L +I+ + LKE WIS QP L V YL QYMPT +W
Sbjct: 251 TKTIGNNGDQ---SHKMTTSRCVRLMLISMANDLKEVWISEQPFLLVTYLWQYMPTWAWW 307
Query: 224 LMDKVGKNRVEAAKEKGNAYS 244
+ +K+GK R+E K +A S
Sbjct: 308 ITNKMGKKRIENFKSGVDADS 328
>DHS7_MOUSE (Q9CXR1) Dehydrogenase/reductase SDR family member 7
precursor (EC 1.1.-.-) (Retinal short-chain
dehydrogenase/reductase 4)
Length = 338
Score = 112 bits (280), Expect = 9e-25
Identities = 65/191 (34%), Positives = 102/191 (53%), Gaps = 13/191 (6%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++N + +S VL+ + K ++N GT++LT+ + P M+ R +G V ++S
Sbjct: 132 IDILVNNGGRSQ-RSLVLETNLDVFKELINLNYIGTVSLTKCVLPHMIERKQGKIVTVNS 190
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQ-KGIQVTVVCPGPI-----------ET 166
AG + Y ASK+AL G+F++L SEL Q GI V PGP+ E
Sbjct: 191 IAGIASVSLSSGYCASKHALRGFFNALHSELGQYPGITFCNVYPGPVQSDIVKNAFTEEV 250
Query: 167 ANNSGSQVPSEKRVSAEKCVELTIIAATHGLKEAWISYQPVLAVMYLVQYMPTIGYWLMD 226
+ + + ++ +CV L +I+ + LKE WIS PVL Y+ QYMPT WL
Sbjct: 251 TKSMRNNIDQSYKMPTSRCVRLMLISMANDLKEVWISDHPVLLGAYIWQYMPTWAAWLNC 310
Query: 227 KVGKNRVEAAK 237
K+GK R++ K
Sbjct: 311 KLGKERIQNFK 321
>YQJQ_BACSU (P54554) Hypothetical oxidoreductase yqjQ (EC 1.-.-.-)
Length = 259
Score = 96.3 bits (238), Expect = 6e-20
Identities = 51/108 (47%), Positives = 73/108 (67%), Gaps = 1/108 (0%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +I+NA + ++ VLD T + +KA FDVNVFG I T+ + P ML + KGH + ++S
Sbjct: 81 IDVLINNAGFGIFET-VLDSTLDDMKAMFDVNVFGLIACTKAVLPQMLEQKKGHIINIAS 139
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
AGK P ++YSA+K+A+ GY ++LR EL GI VT V PGPI+T
Sbjct: 140 QAGKIATPKSSLYSATKHAVLGYSNALRMELSGTGIYVTTVNPGPIQT 187
>DHS4_HUMAN (Q9BTZ2) Dehydrogenase/reductase SDR family member 4 (EC
1.1.1.184) (NADPH-dependent carbonyl
reductase/NADP-retinol dehydrogenase) (CR) (PHCR)
(Peroxisomal short-chain alcohol dehydrogenase)
(NADPH-dependent retinol dehydrogenase/reductase) (
Length = 260
Score = 82.0 bits (201), Expect = 1e-15
Identities = 46/131 (35%), Positives = 74/131 (56%)
Query: 58 GVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMS 117
G+D ++ NAA S++DVTEE T D+NV +T+ + P M +RG G V++S
Sbjct: 91 GIDILVSNAAVNPFFGSIMDVTEEVWDKTLDINVKAPALMTKAVVPEMEKRGGGSVVIVS 150
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQVPSE 177
S A +P+PG + Y+ SK AL G +L EL + I+V + PG I+T+ + + E
Sbjct: 151 SIAAFSPSPGFSPYNVSKTALLGLTKTLAIELAPRNIRVNCLAPGLIKTSFSRMLWMDKE 210
Query: 178 KRVSAEKCVEL 188
K S ++ + +
Sbjct: 211 KEESMKETLRI 221
>YBBO_ECOLI (P77388) Hypothetical oxidoreductase ybbO (EC 1.-.-.-)
Length = 269
Score = 76.6 bits (187), Expect = 5e-14
Identities = 56/165 (33%), Positives = 81/165 (48%), Gaps = 18/165 (10%)
Query: 78 VTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYA 137
++ ++ F N FG LT L P ML G+G V+ SS G PG+ Y+ASKYA
Sbjct: 106 ISRAQMEQQFSANFFGAHQLTMRLLPAMLPHGEGRIVMTSSVMGLISTPGRGAYAASKYA 165
Query: 138 LNGYFHSLRSELCQKGIQVTVVCPGPIET-----ANNSGSQVPSEKRVSAEKCVELTIIA 192
L + +LR EL GI+V+++ PGPI T N + S P E A + L A
Sbjct: 166 LEAWSDALRMELRHSGIKVSLIEPGPIRTRFTDNVNQTQSDKPVENPGIAAR-FTLGPEA 224
Query: 193 ATHGLKEAWISYQPVL---------AVMYLVQYMPTIGYWLMDKV 228
++ A+IS +P + AVM L + +P +MDK+
Sbjct: 225 VVDKVRHAFISEKPKMRYPVTLVTWAVMVLKRLLPG---RVMDKI 266
>DHS4_BOVIN (Q8SPU8) Dehydrogenase/reductase SDR family member 4 (EC
1.1.1.184) (NADPH-dependent carbonyl
reductase/NADP-retinol dehydrogenase) (CR) (PHCR)
(Peroxisomal short-chain alcohol dehydrogenase)
(NADPH-dependent retinol dehydrogenase/reductase) (
Length = 260
Score = 76.6 bits (187), Expect = 5e-14
Identities = 44/110 (40%), Positives = 60/110 (54%)
Query: 58 GVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMS 117
GVD +I NAA S++DV EE DVNV T LT+ + P M +RG G V++S
Sbjct: 91 GVDILISNAAVSPFFGSLMDVPEEVWDKILDVNVKATALLTKAVVPEMAKRGGGSIVIVS 150
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETA 167
S A +P P Y+ SK AL G +L EL + ++V + PG I T+
Sbjct: 151 SIAAYSPFPSLGPYNVSKTALLGLTKNLALELAESNVRVNCLAPGLIRTS 200
>UCPA_SALTY (P37441) Oxidoreductase ucpA (EC 1.-.-.-)
Length = 263
Score = 72.8 bits (177), Expect = 8e-13
Identities = 46/141 (32%), Positives = 69/141 (48%), Gaps = 8/141 (5%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +++NA R + LD++EE D+N+ G +T+ + P M++R G V+MSS
Sbjct: 83 IDILVNNAGVCR-LGNFLDMSEEDRDFHIDINIKGVWNVTKAVLPEMIKRKDGRIVMMSS 141
Query: 119 AAGKTPA-PGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET------ANNSG 171
G A PG+ Y+ SK A+ G SL E Q GI+V +CPG + T A S
Sbjct: 142 VTGDMVADPGETAYALSKAAIVGLTKSLAVEYAQSGIRVNAICPGYVRTPMAESIARQSN 201
Query: 172 SQVPSEKRVSAEKCVELTIIA 192
P K + L +A
Sbjct: 202 PDDPESVLTEMAKAIPLRRLA 222
>HETN_ANASP (P37694) Ketoacyl reductase hetN (EC 1.3.1.-)
Length = 287
Score = 72.4 bits (176), Expect = 1e-12
Identities = 44/126 (34%), Positives = 70/126 (54%), Gaps = 6/126 (4%)
Query: 59 VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMSS 118
+D +I+NA E + + + +++ F+ N+ + LTRLL P M+ RG G V ++S
Sbjct: 84 IDVLINNAGIEI-NGTFANYSLAEIQSIFNTNLLAAMELTRLLLPSMMERGSGRIVNIAS 142
Query: 119 AAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNSGSQVPSEK 178
AGK +VYSASK L + ++R EL G+ ++VVCPG + S + + +
Sbjct: 143 LAGKKGVAFNSVYSASKAGLIMWTDAMRQELVGTGVNISVVCPGYV-----SQTGMTVDT 197
Query: 179 RVSAEK 184
RVSA K
Sbjct: 198 RVSAPK 203
>Y4EK_RHISN (P55434) Putative short-chain type
dehydrogenase/reductase Y4EK (EC 1.-.-.-)
Length = 248
Score = 72.0 bits (175), Expect = 1e-12
Identities = 37/120 (30%), Positives = 64/120 (52%)
Query: 47 VDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFML 106
V E +D +++NA +S++D++E+ + A D+N+ LTR + M+
Sbjct: 63 VSEVYGKIAGEKIDVLVNNAGLLTASASLVDLSEDDIDAMIDINIRSVFKLTRHVLKQMI 122
Query: 107 RRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
R +GH S+ G P P +VY A+K A++ + +LR +L I+VT + PG +ET
Sbjct: 123 ERRRGHIFFTGSSGGLAPHPNSSVYGATKAAVSLFSSALRCDLIGLPIRVTELFPGRVET 182
>UCPA_ECO57 (Q8XBJ4) Oxidoreductase ucpA (EC 1.-.-.-)
Length = 263
Score = 71.6 bits (174), Expect = 2e-12
Identities = 50/163 (30%), Positives = 76/163 (45%), Gaps = 13/163 (7%)
Query: 37 PSKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTIT 96
P+ V A K E E +D +++NA R S LD++++ D+N+ G
Sbjct: 66 PASVAAAIKRAKEKEGR-----IDILVNNAGVCR-LGSFLDMSDDDRDFHIDINIKGVWN 119
Query: 97 LTRLLTPFMLRRGKGHFVVMSSAAGKTPA-PGQAVYSASKYALNGYFHSLRSELCQKGIQ 155
+T+ + P M+ R G V+MSS G A PG+ Y+ +K A+ G SL E Q GI+
Sbjct: 120 VTKAVLPEMIARKDGRIVMMSSVTGDMVADPGETAYALTKAAIVGLTKSLAVEYAQSGIR 179
Query: 156 VTVVCPGPIET------ANNSGSQVPSEKRVSAEKCVELTIIA 192
V +CPG + T A S + P K + L +A
Sbjct: 180 VNAICPGYVRTPMAESIARQSNPEDPESVLTEMAKAIPLRRLA 222
>YUSZ_BACSU (P37959) Hypothetical oxidoreductase yusZ (EC 1.-.-.-)
(ORFA)
Length = 280
Score = 70.9 bits (172), Expect = 3e-12
Identities = 41/130 (31%), Positives = 73/130 (55%), Gaps = 5/130 (3%)
Query: 40 VTAKRKVVDEAESLFPDSGVDYMIHNA--AYERPKSSVLDVTEESLKATFDVNVFGTITL 97
VT ++ +V +++ + +D +++NA AY + DV E + F+ NVFG I +
Sbjct: 62 VTDEQSIVSFGKAVSAYAPIDLLVNNAGTAYG---GFIEDVPMEHFRQQFETNVFGVIHV 118
Query: 98 TRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVT 157
T+ + P++ + G + +SS +G T P + Y +SK+AL G+ SLR EL GI+
Sbjct: 119 TKTVLPYIRKHGGAKIINVSSISGLTGFPALSPYVSSKHALEGFSESLRIELLPFGIETA 178
Query: 158 VVCPGPIETA 167
++ PG +T+
Sbjct: 179 LIEPGSYKTS 188
>UCPA_ECOLI (P37440) Oxidoreductase ucpA (EC 1.-.-.-)
Length = 263
Score = 70.9 bits (172), Expect = 3e-12
Identities = 49/163 (30%), Positives = 76/163 (46%), Gaps = 13/163 (7%)
Query: 37 PSKVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTIT 96
P+ V A K E E +D +++NA R S LD++++ D+N+ G
Sbjct: 66 PASVAAAIKRAKEKEGR-----IDILVNNAGVCR-LGSFLDMSDDDRDFHIDINIKGVWN 119
Query: 97 LTRLLTPFMLRRGKGHFVVMSSAAGKTPA-PGQAVYSASKYALNGYFHSLRSELCQKGIQ 155
+T+ + P M+ R G V+MSS G A PG+ Y+ +K A+ G SL E Q GI+
Sbjct: 120 VTKAVLPEMIARKDGRIVMMSSVTGDMVADPGETAYALTKAAIVGLTKSLAVEYAQSGIR 179
Query: 156 VTVVCPGPIET------ANNSGSQVPSEKRVSAEKCVELTIIA 192
V +CPG + T A S + P K + + +A
Sbjct: 180 VNAICPGYVRTPMAESIARQSNPEDPESVLTEMAKAIPMRRLA 222
>DHS4_MOUSE (Q99LB2) Dehydrogenase/reductase SDR family member 4 (EC
1.1.1.184) (NADPH-dependent carbonyl
reductase/NADP-retinol dehydrogenase) (CR) (PHCR)
(Peroxisomal short-chain alcohol dehydrogenase)
(NADPH-dependent retinol dehydrogenase/reductase) (
Length = 260
Score = 70.9 bits (172), Expect = 3e-12
Identities = 40/113 (35%), Positives = 60/113 (52%)
Query: 58 GVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMS 117
G+D ++ NAA +++DVTEE +NV T + + + P M +RG G V++
Sbjct: 91 GIDILVSNAAVNPFFGNLMDVTEEVWDKVLSINVTATAMMIKAVVPEMEKRGGGSVVIVG 150
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETANNS 170
S AG T P Y+ SK AL G + +EL K I+V + PG I+T +S
Sbjct: 151 SVAGFTRFPSLGPYNVSKTALLGLTKNFAAELAPKNIRVNCLAPGLIKTRFSS 203
>DHS4_RAT (Q8VID1) Dehydrogenase/reductase SDR family member 4 (EC
1.1.1.184) (NADPH-dependent carbonyl
reductase/NADP-retinol dehydrogenase) (CR) (PHCR)
(Peroxisomal short-chain alcohol dehydrogenase)
(NADPH-dependent retinol dehydrogenase/reductase) (ND
Length = 260
Score = 70.5 bits (171), Expect = 4e-12
Identities = 43/128 (33%), Positives = 66/128 (50%), Gaps = 2/128 (1%)
Query: 39 KVTAKRKVVDEAESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLT 98
K + K+V+ A L G+D ++ NAA +++DVTEE +NV + +
Sbjct: 74 KAEDREKLVNMALKLH--QGIDILVSNAAVNPFFGNLMDVTEEVWNKVLSINVTASAMMI 131
Query: 99 RLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTV 158
+ + P M +RG G V++SS AG P Y+ SK AL G + +EL K I+V
Sbjct: 132 KAVVPAMEKRGGGSVVIVSSVAGFVLFPSLGPYNVSKTALLGLTKNFAAELAPKNIRVNC 191
Query: 159 VCPGPIET 166
+ PG I+T
Sbjct: 192 LAPGLIKT 199
>DHS4_PIG (Q8WNV7) Dehydrogenase/reductase SDR family member 4 (EC
1.1.1.184) (NADPH-dependent carbonyl
reductase/NADP-retinol dehydrogenase) (CR) (PHCR)
(Peroxisomal short-chain alcohol dehydrogenase)
(NADPH-dependent retinol dehydrogenase/reductase) (ND
Length = 260
Score = 70.1 bits (170), Expect = 5e-12
Identities = 39/109 (35%), Positives = 58/109 (52%)
Query: 58 GVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVMS 117
GVD ++ NAA +++D TEE VNV T+ +T+ + P M +RG G +++S
Sbjct: 91 GVDILVSNAAVNPFFGNIIDATEEVWDKILHVNVKATVLMTKAVVPEMEKRGGGSVLIVS 150
Query: 118 SAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
S P P Y+ SK AL G +L EL + I+V + PG I+T
Sbjct: 151 SVGAYHPFPNLGPYNVSKTALLGLTKNLAVELAPRNIRVNCLAPGLIKT 199
>DHS4_RABIT (Q9GKX2) Dehydrogenase/reductase SDR family member 4 (EC
1.1.1.184) (NADPH-dependent carbonyl
reductase/NADP-retinol dehydrogenase) (CR) (PHCR)
(Peroxisomal short-chain alcohol dehydrogenase)
(NADPH-dependent retinol dehydrogenase/reductase) (
Length = 260
Score = 69.7 bits (169), Expect = 6e-12
Identities = 41/118 (34%), Positives = 62/118 (51%)
Query: 50 AESLFPDSGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRG 109
A +L G+D ++ NAA ++DVTEE D+NV +T+ + P M +RG
Sbjct: 83 ATALNLHGGIDILVSNAAVNPFFGKLMDVTEEVWDKILDINVKAMALMTKAVVPEMEKRG 142
Query: 110 KGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIETA 167
G V+++S A P G Y+ SK AL G +L EL + I+V + PG I+T+
Sbjct: 143 GGSVVIVASIAAFNPFSGLGPYNVSKTALVGLTKNLALELAAQNIRVNCLAPGLIKTS 200
>YM71_YEAST (Q05016) Hypothetical oxidoreductase in MRPL44-MTF1
intergenic region (EC 1.-.-.-)
Length = 267
Score = 68.9 bits (167), Expect = 1e-11
Identities = 38/128 (29%), Positives = 64/128 (49%), Gaps = 1/128 (0%)
Query: 40 VTAKRKVVDEAESLFPD-SGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLT 98
+T K+ E+L + +D +++NA V + E ++ FD NV I +T
Sbjct: 76 ITQAEKIKPFIENLPQEFKDIDILVNNAGKALGSDRVGQIATEDIQDVFDTNVTALINIT 135
Query: 99 RLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTV 158
+ + P + G V + S AG+ P ++Y ASK+A+ + SLR EL I+V +
Sbjct: 136 QAVLPIFQAKNSGDIVNLGSIAGRDAYPTGSIYCASKFAVGAFTDSLRKELINTKIRVIL 195
Query: 159 VCPGPIET 166
+ PG +ET
Sbjct: 196 IAPGLVET 203
>Y484_MYCTU (Q11150) Putative oxidoreductase Rv0484c/MT0502 (EC
1.-.-.-)
Length = 251
Score = 67.4 bits (163), Expect = 3e-11
Identities = 38/110 (34%), Positives = 59/110 (53%)
Query: 57 SGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVM 116
S VD +++NA + V D E + +D NV GT+ +TR L P ++ G G V +
Sbjct: 75 SRVDVLVNNAGGAKGLQFVADADLEHWRWMWDTNVLGTLRVTRALLPKLIDSGDGLIVTV 134
Query: 117 SSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
+S A G A Y+A+K+A +LR EL K +++T + PG +ET
Sbjct: 135 TSIAAIEVYDGGAGYTAAKHAQGALHRTLRGELLGKPVRLTEIAPGAVET 184
>FABG_THEMA (Q9X248) 3-oxoacyl-[acyl-carrier-protein] reductase (EC
1.1.1.100) (3-ketoacyl-acyl carrier protein reductase)
Length = 246
Score = 67.0 bits (162), Expect = 4e-11
Identities = 44/149 (29%), Positives = 77/149 (51%), Gaps = 4/149 (2%)
Query: 35 PKPSKVTAKRKVVDEAESLFPDSG-VDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFG 93
P VT + ++ + E + G +D +++NA R + ++ + EE A +VN+ G
Sbjct: 58 PYVLNVTDRDQIKEVVEKVVQKYGRIDVLVNNAGITRD-ALLVRMKEEDWDAVINVNLKG 116
Query: 94 TITLTRLLTPFMLRRGKGHFVVMSSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKG 153
+T+++ P+M+++ G V +SS G PGQ Y+ASK + G + EL +
Sbjct: 117 VFNVTQMVVPYMIKQRNGSIVNVSSVVGIYGNPGQTNYAASKAGVIGMTKTWAKELAGRN 176
Query: 154 IQVTVVCPGPIETANNSGSQVPSEKRVSA 182
I+V V PG IET ++P + R +A
Sbjct: 177 IRVNAVAPGFIETPMT--EKLPEKARETA 203
>FAG1_SYNY3 (P73574) 3-oxoacyl-[acyl-carrier-protein] reductase 1
(EC 1.1.1.100) (3-ketoacyl-acyl carrier protein
reductase 1)
Length = 247
Score = 65.9 bits (159), Expect = 9e-11
Identities = 36/110 (32%), Positives = 61/110 (54%), Gaps = 1/110 (0%)
Query: 57 SGVDYMIHNAAYERPKSSVLDVTEESLKATFDVNVFGTITLTRLLTPFMLRRGKGHFVVM 116
S +D +++NA R + +L + E +A D+N+ G T+ ++ ML++ G + +
Sbjct: 83 SRIDVLVNNAGITRD-TLLLRMKLEDWQAVIDLNLTGVFLCTKAVSKLMLKQKSGRIINI 141
Query: 117 SSAAGKTPAPGQAVYSASKYALNGYFHSLRSELCQKGIQVTVVCPGPIET 166
+S AG PGQA YSA+K + G+ ++ EL +G+ V V PG I T
Sbjct: 142 TSVAGMMGNPGQANYSAAKAGVIGFTKTVAKELASRGVTVNAVAPGFIAT 191
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,751,260
Number of Sequences: 164201
Number of extensions: 1087512
Number of successful extensions: 2853
Number of sequences better than 10.0: 314
Number of HSP's better than 10.0 without gapping: 239
Number of HSP's successfully gapped in prelim test: 75
Number of HSP's that attempted gapping in prelim test: 2539
Number of HSP's gapped (non-prelim): 329
length of query: 254
length of database: 59,974,054
effective HSP length: 108
effective length of query: 146
effective length of database: 42,240,346
effective search space: 6167090516
effective search space used: 6167090516
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147179.10


