

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.5 + phase: 0 /pseudo
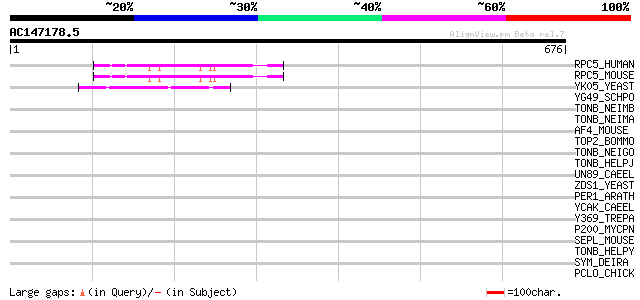
(676 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RPC5_HUMAN (Q9NVU0) DNA-directed RNA polymerases III 80 kDa poly... 89 5e-17
RPC5_MOUSE (Q9CZT4) DNA-directed RNA polymerases III 80 kDa poly... 88 8e-17
YK05_YEAST (P36121) Hypothetical 32.1 kDa protein in DBP7-GCN3 i... 50 1e-05
YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II 39 0.058
TONB_NEIMB (P57004) TonB protein 38 0.076
TONB_NEIMA (P57003) TonB protein 38 0.076
AF4_MOUSE (O88573) AF-4 protein (Proto-oncogene AF4) 38 0.100
TOP2_BOMMO (O16140) DNA topoisomerase II (EC 5.99.1.3) (TOPOII) 37 0.13
TONB_NEIGO (O06432) TonB protein 37 0.13
TONB_HELPJ (Q9ZJP4) TonB protein 37 0.17
UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89 (Uncoo... 37 0.22
ZDS1_YEAST (P50111) ZDS1 protein (NRC1 protein) (RT2GS1) 36 0.29
PER1_ARATH (Q96506) Peroxidase 1/2 precursor (EC 1.11.1.7) (Atpe... 36 0.29
YCAK_CAEEL (Q18563) Hypothetical protein C41G11.3 in chromosome X 35 0.49
Y369_TREPA (O83384) Hypothetical protein TP0369 precursor 35 0.65
P200_MYCPN (P75211) Protein P200 35 0.65
SEPL_MOUSE (Q62170) P-selectin glycoprotein ligand 1 precursor (... 34 1.1
TONB_HELPY (O25899) TonB protein 34 1.4
SYM_DEIRA (Q9RUF3) Methionyl-tRNA synthetase (EC 6.1.1.10) (Meth... 34 1.4
PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment) 34 1.4
>RPC5_HUMAN (Q9NVU0) DNA-directed RNA polymerases III 80 kDa
polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5)
(RPC5)
Length = 708
Score = 88.6 bits (218), Expect = 5e-17
Identities = 80/268 (29%), Positives = 121/268 (44%), Gaps = 57/268 (21%)
Query: 103 DEEDTVVREIDVYFSPSIDNDTKLYVMQYPLRPSWRPYDLDEQCEEVRLKPGTSEVEVDL 162
+E+D VV+EIDVY + S+ KLY+ QYP+RP+ YD D ++KP +VE+++
Sbjct: 4 EEDDPVVQEIDVYLAKSLAE--KLYLFQYPVRPASMTYD-DIPHLSAKIKPKQQKVELEM 60
Query: 163 SIDLES------------YNVEKATADKLK------YTKQTLSTTWKPPPANGCAVGLLM 204
+ID + NV+ A AD+ KQT ++ + A L
Sbjct: 61 AIDTLNPNYCRSKGEQIALNVDGACADETSTYSSKLMDKQTFCSSQTTSNTSRYAAALYR 120
Query: 205 GDKLHLHPIHAVVQLRPSRHYLDSGGS-----EKKNVATSNNQN------KQTT-----P 248
+LHL P+H ++QLRPS YLD + E N A ++Q+ KQ T P
Sbjct: 121 QGELHLTPLHGILQLRPSFSYLDKADAKHREREAANEAGDSSQDEAEDDVKQITVRFSRP 180
Query: 249 SMEQ-KSDGAPSIE--QKSDSNQCWVPLEYHGCKSDISSRYFQQMVTHESSPINFEMSTY 305
EQ + S E QK + + WV L Y+G + S Q ++ SS
Sbjct: 181 ESEQARQRRVQSYEFLQKKHAEEPWVHLHYYGLRDSRSEHERQYLLCPGSS--------- 231
Query: 306 DYIATLCPGVSSNTLAKGPSKRYLLSLP 333
GV + L K PS+ ++ +P
Sbjct: 232 --------GVENTELVKSPSEYLMMLMP 251
>RPC5_MOUSE (Q9CZT4) DNA-directed RNA polymerases III 80 kDa
polypeptide (EC 2.7.7.6) (RNA polymerase III subunit 5)
(RPC5) (Sex-lethal interactor homolog)
Length = 710
Score = 87.8 bits (216), Expect = 8e-17
Identities = 79/268 (29%), Positives = 120/268 (44%), Gaps = 57/268 (21%)
Query: 103 DEEDTVVREIDVYFSPSIDNDTKLYVMQYPLRPSWRPYDLDEQCEEVRLKPGTSEVEVDL 162
+E+D V++EIDVY + S+ KLY+ QYP+RP+ YD D ++KP +VE+++
Sbjct: 4 EEDDPVIQEIDVYLAKSLAE--KLYLFQYPVRPASMTYD-DIPHLSAKIKPKQQKVELEM 60
Query: 163 SIDLES------------YNVEKATADKLK------YTKQTLSTTWKPPPANGCAVGLLM 204
+ID + NV+ A AD+ KQT ++ A L
Sbjct: 61 AIDTLNPNYCRSKGEQIALNVDGACADETSTYSSKLMDKQTFCSSQTTSNTARYAAALYR 120
Query: 205 GDKLHLHPIHAVVQLRPSRHYLDSGGS-----EKKNVATSNNQN------KQTT-----P 248
+LHL P+H ++QLRPS YLD + E N A ++Q+ KQ T P
Sbjct: 121 QGELHLTPLHGILQLRPSFSYLDKADAKHREREAANEAGDSSQDEAEEDVKQITVRFSRP 180
Query: 249 SMEQ-KSDGAPSIE--QKSDSNQCWVPLEYHGCKSDISSRYFQQMVTHESSPINFEMSTY 305
EQ + S E QK + + WV L Y+G + S Q ++ SS
Sbjct: 181 ESEQARQRRVQSYEFLQKKHAEEPWVHLHYYGMRDSRSEHERQYLLCQGSS--------- 231
Query: 306 DYIATLCPGVSSNTLAKGPSKRYLLSLP 333
GV + L K PS+ ++ +P
Sbjct: 232 --------GVENTELVKSPSEYLMMLMP 251
>YK05_YEAST (P36121) Hypothetical 32.1 kDa protein in DBP7-GCN3
intergenic region
Length = 282
Score = 50.4 bits (119), Expect = 1e-05
Identities = 48/188 (25%), Positives = 78/188 (40%), Gaps = 12/188 (6%)
Query: 84 NVDVEMTEAVDGSREVNPMDEEDTVVREIDVYFSPSIDNDTKLYVMQYPLRPSWRPYDLD 143
N E +E V + +E+D V+ E + S + L+V QY RP
Sbjct: 46 NEQEEKSEEVKAEDDTGEEEEDDPVIEEFPLKIS---GEEESLHVFQYANRPRLVGRKPA 102
Query: 144 EQ--CEEVRLKPGTSEVEVDLSIDLES-YNVEKATADKLKYTKQTLSTTWKPPPANGCAV 200
E R KP + E+D+ +D ++ YN +KA ++ QTL NG
Sbjct: 103 EHPFISAARYKPKSHLWEIDIPLDEQAFYNKDKAESEWNGVNVQTLKGVGVEN--NGQYA 160
Query: 201 GLLMGDKLHLHPIHAVVQLRPSRHYLDSGGSEKKNVATSNNQNKQTTPSMEQKSDGAPSI 260
+ +++L PI V QL+P Y+D +K N N PS ++ S+
Sbjct: 161 AFVKDMQVYLVPIERVAQLKPFFKYIDDANVTRKQEDARRNPN----PSSQRAQVVTMSV 216
Query: 261 EQKSDSNQ 268
+ +D +Q
Sbjct: 217 KSVNDPSQ 224
>YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II
Length = 1102
Score = 38.5 bits (88), Expect = 0.058
Identities = 31/123 (25%), Positives = 52/123 (42%), Gaps = 8/123 (6%)
Query: 12 PRVSRFAPKSSKFKPKKEEPSLVPKSEPPPLASAKTEPQEIDLTAPT------PVKHEPN 65
P S + + KP+K EP+L P S K QE + TA T PVK +
Sbjct: 896 PSSSSKSDAAKSIKPQKPEPALKPVEGTADPKSTKRNHQETEKTADTDVSSTEPVKRQKT 955
Query: 66 ASVKTD-AESKIEGKVDSRNVDVEMT-EAVDGSREVNPMDEEDTVVREIDVYFSPSIDND 123
A V D E +++ V + ++T E S +P ++ D ++ + I+ D
Sbjct: 956 ADVNDDVGEEEVKQSVSEQVDSAQLTSEPKSESLPKSPEEKSDDTSNDVTTENTNDINGD 1015
Query: 124 TKL 126
+ +
Sbjct: 1016 SNM 1018
>TONB_NEIMB (P57004) TonB protein
Length = 280
Score = 38.1 bits (87), Expect = 0.076
Identities = 27/72 (37%), Positives = 37/72 (50%), Gaps = 8/72 (11%)
Query: 10 VKPRVSRFAP---KSSKFKPKKEE---PSLVPKSEPPPLASAKTEPQEIDLTAPT--PVK 61
+KP V++ A + K +PK EE P PK EP P A +P E + P+ P +
Sbjct: 84 LKPVVTKKADADIQQPKEEPKPEEKPKPEEKPKPEPKPEAKPVPKPAEKPVEKPSEKPAE 143
Query: 62 HEPNASVKTDAE 73
H NAS K D+E
Sbjct: 144 HPGNASAKADSE 155
>TONB_NEIMA (P57003) TonB protein
Length = 280
Score = 38.1 bits (87), Expect = 0.076
Identities = 27/72 (37%), Positives = 37/72 (50%), Gaps = 8/72 (11%)
Query: 10 VKPRVSRFAP---KSSKFKPKKEE---PSLVPKSEPPPLASAKTEPQEIDLTAPT--PVK 61
+KP V++ A + K +PK EE P PK EP P A +P E + P+ P +
Sbjct: 84 LKPVVTKKADADIQQPKEEPKPEEKPKPEEKPKPEPKPEAKPVPKPAEKPVEKPSEKPAE 143
Query: 62 HEPNASVKTDAE 73
H NAS K D+E
Sbjct: 144 HPGNASAKADSE 155
>AF4_MOUSE (O88573) AF-4 protein (Proto-oncogene AF4)
Length = 1217
Score = 37.7 bits (86), Expect = 0.100
Identities = 50/201 (24%), Positives = 75/201 (36%), Gaps = 31/201 (15%)
Query: 5 DLDAPVKPRVSRFA-PKSSKFKPKKEEPSLVPK-------SEPPPLASAKTEPQEIDLTA 56
DL P K +++R P S +P E V + S PPPL + T
Sbjct: 312 DLKVPAKAKLTRLRMPSQSVEQPYSNEVHCVEEILKEMTHSWPPPLTAIHTPSTAEPSRF 371
Query: 57 PTPVKHEPNASVKTDAESKIEGKVDSRNVDVEMTEAVDGSREVNPMDEEDTVVREIDVYF 116
P P K + S T ++ + D + +P ++ T + E D+
Sbjct: 372 PFPTKDLLHVSPATQSQKQ-----------------YDTPSKTHPNPQQGTSMLEDDLQL 414
Query: 117 SPSIDNDTKLYVMQYPLRPS--WRPYDLDEQCEEVRLKPGTSE-VEVDLSIDLESYNVEK 173
S S D+DT+ + P P+ P L E G SE E D S D ES E
Sbjct: 415 SDSEDSDTEQATEKPPSPPAPPSAPQTLPEPVASAHSSSGESESSESDSSSDSES---ES 471
Query: 174 ATADKLKYTKQTLSTTWKPPP 194
+++D + + T P P
Sbjct: 472 SSSDSEEEEENEPLETRAPEP 492
>TOP2_BOMMO (O16140) DNA topoisomerase II (EC 5.99.1.3) (TOPOII)
Length = 1547
Score = 37.4 bits (85), Expect = 0.13
Identities = 62/281 (22%), Positives = 110/281 (39%), Gaps = 30/281 (10%)
Query: 5 DLDAP--VKPRVSRFAPKS--SKFKPKK-EEPSLVPKSEPPP-----LASAKTEPQEIDL 54
D D P + P PK+ K KP+K E+P V K+E + K EP++ +
Sbjct: 1271 DPDDPTGISPSSGEKKPKARVKKEKPEKAEKPDKVDKAEKTDGLKQTKLTFKKEPKKKKM 1330
Query: 55 TAPTPVKHEPNASVKTDAESKI---EGKVDSRNVDVEMTEAVDGSREVNPMDEEDTVVRE 111
T E +AS DAE ++ + ++R + + DGS + E + + +
Sbjct: 1331 TFSGSSSGEMSAS---DAEVEVLVPRERTNARRAATRVQKYKDGSDDSGSDSEPELLDNK 1387
Query: 112 IDV-YFSPSI----DNDTKLYVMQYPLRPSWRPYDLDEQCEEVRLKPGTSEVEVDLSIDL 166
ID + +P D D + + P + +P ++D C L + E + L
Sbjct: 1388 IDSDHEAPQTLSISDEDDDFNIKKNPAK---KPAEMDSDCLFDSLIEDAKKDEPQKT--L 1442
Query: 167 ESYNVEKATADKLKYTKQTLSTTWKPPPANGCAVGLLMGDKLHLHPIHAVVQLRPSRHYL 226
E +L +Q T PP LM ++ + RP+R L
Sbjct: 1443 TKSKSESILIRRLNIERQRRCDTSVPPKEKAAPKRKLM----NVDKDEKKTKKRPARVML 1498
Query: 227 DSGGSEKKNVATSNNQNKQTTPSMEQKSDGAPSIEQKSDSN 267
D ++ ++ S K+T + ++K+ + +SD N
Sbjct: 1499 DQDSDDEDSIFDSKKGKKKTAANPKKKAKKKVESDSESDFN 1539
>TONB_NEIGO (O06432) TonB protein
Length = 283
Score = 37.4 bits (85), Expect = 0.13
Identities = 22/66 (33%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query: 10 VKPRVSRFAPKSSKFKPKKEEPSLVPKSEPPPLASAKTEPQEIDLTAPT--PVKHEPNAS 67
+KP V++ A + +K +P PK EP P A +P E P+ P +H NAS
Sbjct: 84 LKPAVTKKADADIQQPKEKPKPEEKPKPEPEPEAKPAPKPAEKPAEKPSEKPAEHSGNAS 143
Query: 68 VKTDAE 73
K +E
Sbjct: 144 AKAGSE 149
>TONB_HELPJ (Q9ZJP4) TonB protein
Length = 280
Score = 37.0 bits (84), Expect = 0.17
Identities = 29/115 (25%), Positives = 45/115 (38%), Gaps = 14/115 (12%)
Query: 20 KSSKFKPKKEEPSL----VPKSEPPPLASAKTEPQEIDLTAPTPVKHEPNASVKTDAESK 75
K K +PKKEEP PK +P P K +P+ P P EP + K
Sbjct: 80 KPKKEEPKKEEPKKETKPKPKPKPKPKPKPKPKPKPEPKPKPEPKVEEPKKEEPKEEPKK 139
Query: 76 IEGKVDSRNVDV-------EMTEAVDGSREVNPMDEEDTVVREIDVYFSPSIDND 123
E K +++ V ++ + D E N E T + ++P + N+
Sbjct: 140 EEAKEEAKEKSVPKQVTTKDIVKEKDKQEESNKTSEGATSEAQA---YNPGVSNE 191
>UN89_CAEEL (O01761) Muscle M-line assembly protein unc-89
(Uncoordinated protein 89)
Length = 6632
Score = 36.6 bits (83), Expect = 0.22
Identities = 54/264 (20%), Positives = 101/264 (37%), Gaps = 22/264 (8%)
Query: 4 DDLDAPVK--PRVSRFAPKSSKFKPKKEEPSLVPKSEPPPLASAKTEPQEIDLTAPTPVK 61
++L +P + P + PKS K K E S + P + P++++ +P K
Sbjct: 1614 EELKSPKEKSPEKADDKPKSPTKKEKSPEKSATEDVKSP--TKKEKSPEKVEEKPTSPTK 1671
Query: 62 HEPNASVKTDAESKIEGKVDSRNVDVEMTEAVDGSREVNPMDEEDTVVREIDVYFSPSID 121
E + + KTD E K K + VE A +E +P E +VV E+ SP
Sbjct: 1672 KESSPTKKTDDEVKSPTKKEKSPQTVEEKPASPTKKEKSP---EKSVVEEVK---SPKEK 1725
Query: 122 NDTKLYVMQYPLRPSWRPYDLDEQCEEVRLKPGTSEVEVDLSIDLESYNVEKATADKLKY 181
+ K + P P+ + ++ E P E + S + + + K + +K
Sbjct: 1726 SPEK--AEEKPKSPTKKEKSPEKSAAEEVKSPTKKEKSPEKSAEEKPKSPTKKESSPVKM 1783
Query: 182 TKQTLSTTWKPPPANGCAVGLLMGDKLHLHPIHAVVQLR-PSRHYLDSGGSEKKNVATSN 240
+ + K + +K+ P + + P + + S K + +
Sbjct: 1784 ADDEVKSPTKKEKS---------PEKVEEKPASPTKKEKTPEKSAAEELKSPTKKEKSPS 1834
Query: 241 NQNKQTTPSMEQKSDGAPSIEQKS 264
+ K+T ++KS P + KS
Sbjct: 1835 SPTKKTGDESKEKSPEKPEEKPKS 1858
>ZDS1_YEAST (P50111) ZDS1 protein (NRC1 protein) (RT2GS1)
Length = 915
Score = 36.2 bits (82), Expect = 0.29
Identities = 21/72 (29%), Positives = 38/72 (52%), Gaps = 3/72 (4%)
Query: 204 MGDKLHLHPIHAVV---QLRPSRHYLDSGGSEKKNVATSNNQNKQTTPSMEQKSDGAPSI 260
+G+K P+ +V + RP RH+ GS+K +V T + Q ++ + +GA I
Sbjct: 626 LGNKSGREPVEPIVLRNRPRPHRHHHSRHGSQKISVKTLKDSQPQQQIPLQPQLEGAIEI 685
Query: 261 EQKSDSNQCWVP 272
E+K +S+ +P
Sbjct: 686 EKKEESDSESLP 697
>PER1_ARATH (Q96506) Peroxidase 1/2 precursor (EC 1.11.1.7) (Atperox
P1/P2) (ATP11a)
Length = 325
Score = 36.2 bits (82), Expect = 0.29
Identities = 23/56 (41%), Positives = 30/56 (53%), Gaps = 11/56 (19%)
Query: 614 CDIHGC---YVLKSAQNDPFRDVVIDMLRGSGPNGKLKKAEILEAARRKLGRDVPN 666
C + GC +LKSA+ND RD V PN LK E+++AA+ L R PN
Sbjct: 68 CFVRGCDGSVLLKSAKNDAERDAV--------PNLTLKGYEVVDAAKTALERKCPN 115
>YCAK_CAEEL (Q18563) Hypothetical protein C41G11.3 in chromosome X
Length = 737
Score = 35.4 bits (80), Expect = 0.49
Identities = 25/78 (32%), Positives = 37/78 (47%), Gaps = 11/78 (14%)
Query: 4 DDLDAPVKPRVSRFAPKSSK--FKPKKEEPSLVPKSEPPPLASAKTEPQEIDLTAPTPVK 61
+++ P K R+S F K+ K KP+ E+ L P+S+ P S T P V+
Sbjct: 551 NNVSEPSKNRLSIFKSKTEKKVVKPESEKEKLKPRSDDIPTTSTST---------PMAVE 601
Query: 62 HEPNASVKTDAESKIEGK 79
+PN S T+ K GK
Sbjct: 602 TKPNGSRTTEEPLKRMGK 619
>Y369_TREPA (O83384) Hypothetical protein TP0369 precursor
Length = 516
Score = 35.0 bits (79), Expect = 0.65
Identities = 22/62 (35%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query: 8 APVKPRVSR-----FAPKSSKFKPKKEEPSLVPKSEPPPLASAKTEPQEIDLTAPTPVKH 62
AP PR R +P +SK K + PS P SEPP A + EP+ + + V
Sbjct: 129 APTAPRPHRPSPPPVSPSASKPKQRAVPPSPPPASEPPREAEVQAEPEPAEDSPRAMVPE 188
Query: 63 EP 64
EP
Sbjct: 189 EP 190
Score = 31.2 bits (69), Expect = 9.3
Identities = 67/326 (20%), Positives = 116/326 (35%), Gaps = 56/326 (17%)
Query: 6 LDAPVKPRVSRFAPKSSKFKPKKEEPSLVP------------KSEPPPLASAKTEPQEID 53
L +PVKPR + P P+ P + P + PPP++ + ++P++
Sbjct: 96 LASPVKPREAGSVPDVLPV-PEVSSPHVAPPAPPAPTAPRPHRPSPPPVSPSASKPKQRA 154
Query: 54 LTAPTPVKHEPNASVKTDAESKIEGKVDSRNVDVEMTEAVDGSREVNPMDEEDTVVREID 113
+ P EP + AE E DS V E P DE V R +
Sbjct: 155 VPPSPPPASEPPREAEVQAEP--EPAEDSPRAMVP---------EEPPEDEVPRVSRAVQ 203
Query: 114 VYFSPSID----NDTKLYVMQYPLRPSWRPYDLDEQCEEVRLKPGTSEVEVDLSIDLESY 169
+ ++ + +YV ++ +P R Y + E L ++E E D + +
Sbjct: 204 LAVGQKLEVLYPGEGWVYVGEHTAQPGLR-YHQRKLEESHSLFTFSAEREGDFVLAFSYF 262
Query: 170 N------VEKATADKLKYTKQTLSTTWKPPPANGCAVGLLMGDKLHLHPIHAVVQLRPS- 222
+ V A A K+ ++ L+ + P + P V +L P+
Sbjct: 263 DVFRGDFVSDALAVKVVPKREGLARVVRAPEYR---------RTVSSPPDTVVSELSPAG 313
Query: 223 ----RHYLDSGGSEKKNVATSN----NQNKQTTPSMEQKSDGAPSIEQKSDSNQC---WV 271
R +SG S + A Q++ T E+ G P ++K D
Sbjct: 314 TGTERRAEESGTSGSQRAAAHTGAPVRQDQTDTAVAEKAQHGTPRPDEKKDREPTVGGRD 373
Query: 272 PLEYHGCKSDISSRYFQQMVTHESSP 297
P+ +S RY + ++ S P
Sbjct: 374 PVPSDAVAQGVSERYSPRKISPASQP 399
>P200_MYCPN (P75211) Protein P200
Length = 1036
Score = 35.0 bits (79), Expect = 0.65
Identities = 33/131 (25%), Positives = 54/131 (41%), Gaps = 23/131 (17%)
Query: 5 DLDAPVKPRVSRFAPKSSKFKPKKE-EPSLVP------------------KSEPPPLASA 45
DL+ +P F+P+ E EP + P +SEP P
Sbjct: 724 DLETIPEPESIETTEPEPNFEPEVELEPEIEPNFESETEVQQELAQESSFESEPEPNFET 783
Query: 46 KTEPQ---EIDLTAPTPVKHEPNASVKTDAESKIEGKVDSRNVDVEMTEAVDGSREVNPM 102
+ E Q EI+ V+ EP S+ +D E+K E + + +E+ EA + E+ P
Sbjct: 784 EVEVQPESEIESKFEAEVQSEPKVSLNSDFETKPEAQAEVTPETLEV-EATSEAPELQPE 842
Query: 103 DEEDTVVREID 113
E VV +++
Sbjct: 843 TEATKVVDDVE 853
>SEPL_MOUSE (Q62170) P-selectin glycoprotein ligand 1 precursor
(PSGL-1) (Selectin P ligand)
Length = 397
Score = 34.3 bits (77), Expect = 1.1
Identities = 25/124 (20%), Positives = 47/124 (37%), Gaps = 11/124 (8%)
Query: 8 APVKPRVSRFAPKSSKFKPKKEEPSLVPKSEPPPLASAKTEPQEIDLTAPTPVKHEPNAS 67
AP++ S+ AP ++ + K P ++K P E + + P P + E +
Sbjct: 148 APMEAETSQPAPMEAETSQPAPMEADTSKPAPTEAETSKPAPTEAETSQPAPNEAETSKP 207
Query: 68 VKTDAESKIEGKVDSRNVDVEMTEAVD-----------GSREVNPMDEEDTVVREIDVYF 116
T+AE+ ++ + +AV S E M+ T E ++
Sbjct: 208 APTEAETSKPAPTEAETTQLPRIQAVKTLFTTSAATEVPSTEPTTMETASTESNESTIFL 267
Query: 117 SPSI 120
PS+
Sbjct: 268 GPSV 271
>TONB_HELPY (O25899) TonB protein
Length = 285
Score = 33.9 bits (76), Expect = 1.4
Identities = 35/121 (28%), Positives = 46/121 (37%), Gaps = 21/121 (17%)
Query: 20 KSSKFKPKKEEPSLV-------PKSEPPPLASAKTEPQEIDLTAPTP-------VKHEPN 65
K K +PKKEEP PK +P P K EP+ P P K EP
Sbjct: 80 KPKKEEPKKEEPKKEVTKPKPKPKPKPKPKPKPKPEPKPEPKPEPKPEPKVEEVKKEEPK 139
Query: 66 ASVKTDAESKIEGKVDSRNVDVEMTEAV---DGSREVNPMDEEDTVVREIDVYFSPSIDN 122
K + E+K E K S V + V D E N E T + ++P + N
Sbjct: 140 EEPKKE-EAKEEAKEKSAPKQVTTKDIVKEKDKQEESNKTSEGATSEAQA---YNPGVSN 195
Query: 123 D 123
+
Sbjct: 196 E 196
Score = 32.3 bits (72), Expect = 4.2
Identities = 29/98 (29%), Positives = 41/98 (41%), Gaps = 12/98 (12%)
Query: 23 KFKPKKEEPSL------VPKSEPPPLASAKTEPQEIDLTAPTPVKHEPNASVKTDAESKI 76
K KPKKEEP V K +P P K +P+ P P K EP K + K
Sbjct: 78 KEKPKKEEPKKEEPKKEVTKPKPKPKPKPKPKPKPKPEPKPEP-KPEPKPEPKVEEVKKE 136
Query: 77 EGKVDSRNVDV-EMTEAVDGSREVNPMDEEDTVVREID 113
E K + + + E + ++V D +V+E D
Sbjct: 137 EPKEEPKKEEAKEEAKEKSAPKQVTTKD----IVKEKD 170
>SYM_DEIRA (Q9RUF3) Methionyl-tRNA synthetase (EC 6.1.1.10)
(Methionine--tRNA ligase) (MetRS)
Length = 681
Score = 33.9 bits (76), Expect = 1.4
Identities = 36/120 (30%), Positives = 51/120 (42%), Gaps = 12/120 (10%)
Query: 9 PVKPRVSRFAPKSSKFKPKKEEPSLVPKSEPPPLASAKTEPQEIDLTAP---TPVKHEPN 65
P RV A K +PK +E P P AK E + + TAP T K E
Sbjct: 510 PAGTRVQGGAILFPKPEPKADETKNAEAKPPKP--QAKKEKKTVTDTAPAKTTEQKPEAA 567
Query: 66 ASVKTDAESKIE--GKVDSRNVDVEMTEAVDGSREVNPM-----DEEDTVVREIDVYFSP 118
A + D I+ K+D R +V EAV+ + ++ + DE TVV I ++ P
Sbjct: 568 APAQNDGLISIDDFAKIDLRIAEVVACEAVEKADKLLKLTVKLGDETRTVVSGIRKWYEP 627
>PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment)
Length = 5120
Score = 33.9 bits (76), Expect = 1.4
Identities = 44/189 (23%), Positives = 76/189 (39%), Gaps = 26/189 (13%)
Query: 12 PRVSRFAPKSSKFKPKKEE--PSLVPKSEPPPLASAKTEPQEIDLTAPTPVKHEPNASVK 69
P+ S P + KPK+E P VPK + L + + K + K
Sbjct: 932 PQKSSTPPTPAATKPKEEPSVPKEVPKLQQGKLEKTLSADKIQQGIQKEDAKSKQGKLFK 991
Query: 70 TDAESKIEG---KVDSRNVDVEMTEAVDGSREVNPMDEEDTVVREIDVYFSPSID----- 121
T + KI+ K DSR ++T+ + ++ + +ED +E + PS D
Sbjct: 992 TPSADKIQRVSQKEDSRLQQTKLTKTPSSDKILHGVQKEDIKFQEAKLAKIPSADKILHR 1051
Query: 122 ---NDTKLYVMQY-------PLRPSWRPYDLDEQCEEVRLKPGTSEVEVDLSI----DLE 167
D KL M+ ++P + D+ Q +EVRL S ++ I +L+
Sbjct: 1052 LQKEDPKLQQMKMAKALSADKIQPEAQKEDV--QLQEVRLSKAVSADKIQHGIQKDLNLQ 1109
Query: 168 SYNVEKATA 176
+EK ++
Sbjct: 1110 HVKIEKTSS 1118
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.327 0.140 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,701,128
Number of Sequences: 164201
Number of extensions: 3119549
Number of successful extensions: 12482
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 12268
Number of HSP's gapped (non-prelim): 231
length of query: 676
length of database: 59,974,054
effective HSP length: 117
effective length of query: 559
effective length of database: 40,762,537
effective search space: 22786258183
effective search space used: 22786258183
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147178.5


