

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147178.4 - phase: 0
(248 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
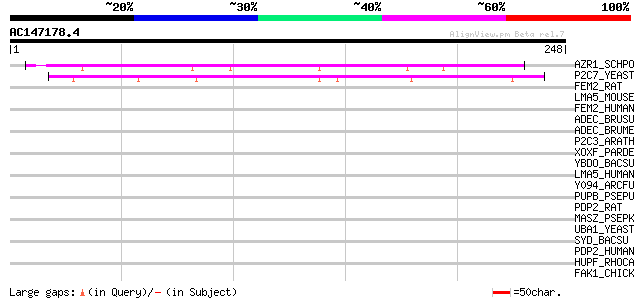
AZR1_SCHPO (Q09189) 5-azacytidine resistance protein azr1 127 3e-29
P2C7_YEAST (P38797) Protein phosphatase 2C homolog 7 (EC 3.1.3.1... 89 1e-17
FEM2_RAT (Q9WVR7) Ca(2+)/calmodulin-dependent protein kinase pho... 41 0.002
LMA5_MOUSE (Q61001) Laminin alpha-5 chain precursor 39 0.015
FEM2_HUMAN (P49593) Ca(2+)/calmodulin-dependent protein kinase p... 39 0.015
ADEC_BRUSU (Q8FW12) Adenine deaminase (EC 3.5.4.2) (Adenase) (Ad... 35 0.22
ADEC_BRUME (Q8YCA5) Adenine deaminase (EC 3.5.4.2) (Adenase) (Ad... 34 0.29
P2C3_ARATH (P49599) Protein phosphatase 2C PPH1 (EC 3.1.3.16) (P... 34 0.38
XOXF_PARDE (P29968) Putative dehydrogenase XOXF precursor (EC 1.... 33 0.84
YBDO_BACSU (O31437) Hypothetical protein ybdO 31 3.2
LMA5_HUMAN (O15230) Laminin alpha-5 chain precursor 30 5.4
Y094_ARCFU (O30142) Hypothetical UPF0100 protein AF0094 30 7.1
PUPB_PSEPU (P38047) Ferric-pseudobactin BN7/BN8 receptor precursor 30 7.1
PDP2_RAT (O88484) [Pyruvate dehydrogenase [Lipoamide]]-phosphata... 30 7.1
MASZ_PSEPK (Q88QX8) Malate synthase G (EC 2.3.3.9) 30 7.1
UBA1_YEAST (P22515) Ubiquitin-activating enzyme E1 1 29 9.3
SYD_BACSU (O32038) Aspartyl-tRNA synthetase (EC 6.1.1.12) (Aspar... 29 9.3
PDP2_HUMAN (Q9P2J9) [Pyruvate dehydrogenase [Lipoamide]]-phospha... 29 9.3
HUPF_RHOCA (Q03005) Hydrogenase expression/formation protein hupF 29 9.3
FAK1_CHICK (Q00944) Focal adhesion kinase 1 (EC 2.7.1.112) (FADK... 29 9.3
>AZR1_SCHPO (Q09189) 5-azacytidine resistance protein azr1
Length = 288
Score = 127 bits (319), Expect = 3e-29
Identities = 83/242 (34%), Positives = 126/242 (51%), Gaps = 23/242 (9%)
Query: 8 LPHPDKVATGGEDAHFICEDEQAI--GVADGVGGWADVGVNAGLYAQELVANSARAIREE 65
L HPD GEDA +E I V DGVGGWA+VG++ +++ LV +
Sbjct: 44 LDHPD----AGEDAFINLRNENYILNAVFDGVGGWANVGIDPSIFSWGLVREIKKVFNNS 99
Query: 66 PKGSFNPVRVLEKAH-----SKTKAMGSSTVCIIALI--DEALNAINLGDSGFIVIRDGS 118
+ +P+ +L KA+ S T GSST C+ + L+++NLGDSGF+++R+G+
Sbjct: 100 DEFQPSPLTLLSKAYAALKKSNTVEAGSSTACLTLFNCGNGKLHSLNLGDSGFLILRNGA 159
Query: 119 VIFKSPVQQRGFNFPYQLA-----RSGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNM 173
+ + SP Q FN PYQLA E P G+ + D+++ TDG+FDN+
Sbjct: 160 IHYASPAQVLQFNMPYQLAIYPRNYRSAENIGPKMGQATVHDLKDNDLVILATDGIFDNI 219
Query: 174 YNN---DIVGVVVGATRARLGP--QATAQKIAALARQRALDTKRQSPFSAAALEYGYRFD 228
DI GVV ++ + + A +I A +LDTK +SPF+ A +G++F
Sbjct: 220 EEKSILDIAGVVDFSSLSNVQKCLDDLAMRICRQAVLNSLDTKWESPFAKTAKSFGFKFQ 279
Query: 229 GG 230
GG
Sbjct: 280 GG 281
>P2C7_YEAST (P38797) Protein phosphatase 2C homolog 7 (EC 3.1.3.16)
(PP2C-7)
Length = 374
Score = 88.6 bits (218), Expect = 1e-17
Identities = 74/251 (29%), Positives = 118/251 (46%), Gaps = 29/251 (11%)
Query: 18 GEDAHFICED---EQAIGVADGVGGWADVGVNAGLYAQELVA-----NSARAIREEPKGS 69
GED +F+ + + GVADGVGGWA+ G ++ ++EL ++A A +
Sbjct: 120 GEDNYFVTSNNVHDIFAGVADGVGGWAEHGYDSSAISRELCKKMDEISTALAENSSKETL 179
Query: 70 FNPVRVLEKAHSK------TKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFKS 123
P +++ A++K K G++ + + L NLGDS V RD ++F++
Sbjct: 180 LTPKKIIGAAYAKIRDEKVVKVGGTTAIVAHFPSNGKLEVANLGDSWCGVFRDSKLVFQT 239
Query: 124 PVQQRGFNFPYQLA-----------RSGTEGDL--PSSGEVFTVPVAPGDIIVAGTDGLF 170
Q GFN PYQL+ R G++ L P + ++ + DII+ TDG+
Sbjct: 240 KFQTVGFNAPYQLSIIPEEMLKEAERRGSKYILNTPRDADEYSFQLKKKDIIILATDGVT 299
Query: 171 DNMYNNDI-VGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEY-GYRFD 228
DN+ +DI + + A R Q +QK + D S F+ + G +
Sbjct: 300 DNIATDDIELFLKDNAARTNDELQLLSQKFVDNVVSLSKDPNYPSVFAQEISKLTGKNYS 359
Query: 229 GGKLDDLTVVV 239
GGK DD+TVVV
Sbjct: 360 GGKEDDITVVV 370
>FEM2_RAT (Q9WVR7) Ca(2+)/calmodulin-dependent protein kinase
phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase)
(CaMKPase) (Partner of PIX 2) (Protein phosphatase 1F)
Length = 450
Score = 41.2 bits (95), Expect = 0.002
Identities = 56/209 (26%), Positives = 91/209 (42%), Gaps = 39/209 (18%)
Query: 33 VADGVGG-----WADVGVNAGLYAQ-ELVANSARAIREEPKGSFNPVRVLEKAHSKTKAM 86
V DG GG +A V V+ Q EL+ + A A++E + + L+KA +
Sbjct: 192 VFDGHGGVDAARYASVHVHTNASHQPELLTDPAAALKEAFRHTDQ--MFLQKAKRERLQS 249
Query: 87 GSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIF------------KSPVQQRGFNFPY 134
G++ VC + + AL+ LGDS I+++ G V+ KS ++ G F
Sbjct: 250 GTTGVCAL-ITGAALHVAWLGDSQVILVQQGQVVKLMEPHKPERQDEKSRIEALG-GFVS 307
Query: 135 QLARSGTEGDLPSS---GEVFTVPVAPG-------------DIIVAGTDGLFDNMYNNDI 178
+ G L S G+VF P G D ++ DG FD + +++I
Sbjct: 308 LMDCWRVNGTLAVSRAIGDVFQKPYVSGEADAASRELTGLEDYLLLACDGFFDVVPHHEI 367
Query: 179 VGVVVG-ATRARLGPQATAQKIAALARQR 206
G+V G R + A+++ A+AR R
Sbjct: 368 PGLVHGHLLRQKGSGMHVAEELVAVARDR 396
>LMA5_MOUSE (Q61001) Laminin alpha-5 chain precursor
Length = 3718
Score = 38.5 bits (88), Expect = 0.015
Identities = 29/92 (31%), Positives = 43/92 (46%), Gaps = 5/92 (5%)
Query: 143 GDLPSSGEVFTVPVAPGDIIVA---GTDGLFDNMYNNDIVGVVVGATRARL--GPQATAQ 197
G LP SG ++ V G +FD N V V VG T A+L +ATAQ
Sbjct: 3251 GGLPVSGTFHNFSGCISNVFVQRLRGPQRVFDLHQNMGSVNVSVGCTPAQLIETSRATAQ 3310
Query: 198 KIAALARQRALDTKRQSPFSAAALEYGYRFDG 229
K++ +RQ + D +P+ ++ Y+F G
Sbjct: 3311 KVSRRSRQPSQDLACTTPWLPGTIQDAYQFGG 3342
>FEM2_HUMAN (P49593) Ca(2+)/calmodulin-dependent protein kinase
phosphatase (EC 3.1.3.16) (CaM-kinase phosphatase)
(CaMKPase) (Partner of PIX 2) (hFEM-2) (Protein
phosphatase 1F)
Length = 454
Score = 38.5 bits (88), Expect = 0.015
Identities = 55/212 (25%), Positives = 88/212 (40%), Gaps = 45/212 (21%)
Query: 33 VADGVGGWADVGVNAGLYAQELV-ANSARA--IREEPKGSFNPV------RVLEKAHSKT 83
V DG GG V+A YA V N+AR + +P+G+ L KA +
Sbjct: 196 VFDGHGG-----VDAARYAAVHVHTNAARQPELPTDPEGALREAFRRTDQMFLRKAKRER 250
Query: 84 KAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIF------------KSPVQQRGFN 131
G++ VC + + L+ LGDS I+++ G V+ K+ ++ G
Sbjct: 251 LQSGTTGVCAL-IAGATLHVAWLGDSQVILVQQGQVVKLMEPHRPERQDEKARIEALG-G 308
Query: 132 FPYQLARSGTEGDLPSS---GEVFTVPVAPG-------------DIIVAGTDGLFDNMYN 175
F + G L S G+VF P G D ++ DG FD + +
Sbjct: 309 FVSHMDCWRVNGTLAVSRAIGDVFQKPYVSGEADAASRALTGSEDYLLLACDGFFDVVPH 368
Query: 176 NDIVGVVVG-ATRARLGPQATAQKIAALARQR 206
++VG+V TR + A+++ A AR+R
Sbjct: 369 QEVVGLVQSHLTRQQGSGLRVAEELVAAARER 400
>ADEC_BRUSU (Q8FW12) Adenine deaminase (EC 3.5.4.2) (Adenase)
(Adenine aminase)
Length = 581
Score = 34.7 bits (78), Expect = 0.22
Identities = 20/66 (30%), Positives = 33/66 (49%), Gaps = 6/66 (9%)
Query: 79 AHSKTKAMGSSTVCIIALIDE----ALNAINLGDSGFIVIRDGSVIFKSPVQQRGF--NF 132
A + T + S +C++ DE A N + + GF+V+RDG V+ + P+ G
Sbjct: 463 AIASTVSHDSHNICVVGASDEDIATAANRLGEIEGGFVVVRDGKVLAEMPLPIAGLMSTE 522
Query: 133 PYQLAR 138
PY+ R
Sbjct: 523 PYETVR 528
>ADEC_BRUME (Q8YCA5) Adenine deaminase (EC 3.5.4.2) (Adenase)
(Adenine aminase)
Length = 581
Score = 34.3 bits (77), Expect = 0.29
Identities = 20/66 (30%), Positives = 33/66 (49%), Gaps = 6/66 (9%)
Query: 79 AHSKTKAMGSSTVCIIALIDE----ALNAINLGDSGFIVIRDGSVIFKSPVQQRGF--NF 132
A + T + S +C++ DE A N + + GF+V+RDG V+ + P+ G
Sbjct: 463 AIASTVSHDSHNICVVGASDEDIATAANRLGEIEGGFVVVRDGKVLAEIPLPIAGLMSTE 522
Query: 133 PYQLAR 138
PY+ R
Sbjct: 523 PYETVR 528
>P2C3_ARATH (P49599) Protein phosphatase 2C PPH1 (EC 3.1.3.16)
(PP2C)
Length = 388
Score = 33.9 bits (76), Expect = 0.38
Identities = 25/91 (27%), Positives = 45/91 (48%), Gaps = 6/91 (6%)
Query: 136 LARSGTEGDLP-SSGEVFTVPVAPG-DIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQ 193
++R +GD+ ++ ++F VP+ + I+ +DGL+D M ++D+V V R Q
Sbjct: 263 VSRIEFKGDMVVATPDIFQVPLTSDVEFIILASDGLWDYMKSSDVVSYVRDQLRKHGNVQ 322
Query: 194 ATAQKIAALARQRALDTKRQSPFSAAALEYG 224
+ +A Q ALD + Q S + G
Sbjct: 323 LACESLA----QVALDRRSQDNISIIIADLG 349
>XOXF_PARDE (P29968) Putative dehydrogenase XOXF precursor (EC
1.1.99.-)
Length = 600
Score = 32.7 bits (73), Expect = 0.84
Identities = 12/22 (54%), Positives = 17/22 (76%)
Query: 28 EQAIGVADGVGGWADVGVNAGL 49
+Q +G+ GVGGWA +G+ AGL
Sbjct: 546 KQYVGILSGVGGWAGIGLAAGL 567
>YBDO_BACSU (O31437) Hypothetical protein ybdO
Length = 394
Score = 30.8 bits (68), Expect = 3.2
Identities = 17/45 (37%), Positives = 27/45 (59%), Gaps = 2/45 (4%)
Query: 45 VNAGLYAQELVANSARAIREEPKGSFNPVRVLEKAHSKTKAMGSS 89
+ L AQ +++ +A A RE + NP VLEKA+S+ K+ S+
Sbjct: 8 IRTQLIAQNILSKNAPAKREN--ATENPAAVLEKAYSRLKSQSST 50
>LMA5_HUMAN (O15230) Laminin alpha-5 chain precursor
Length = 3695
Score = 30.0 bits (66), Expect = 5.4
Identities = 31/108 (28%), Positives = 40/108 (36%), Gaps = 12/108 (11%)
Query: 143 GDLPSSGEVFTVPVAPGDIIVA---GTDGLFDNMYNNDIVGVVVGATRAR------LGP- 192
G LP SG ++ ++ V G +FD N V V G A LGP
Sbjct: 3247 GGLPESGTIYNFSGCISNVFVQRLLGPQRVFDLQQNLGSVNVSTGCAPALQAQTPGLGPR 3306
Query: 193 --QATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVV 238
QATA+K + +RQ A P Y+F G L V
Sbjct: 3307 GLQATARKASRRSRQPARHPACMLPPHLRTTRDSYQFGGSLSSHLEFV 3354
>Y094_ARCFU (O30142) Hypothetical UPF0100 protein AF0094
Length = 342
Score = 29.6 bits (65), Expect = 7.1
Identities = 24/92 (26%), Positives = 41/92 (44%), Gaps = 10/92 (10%)
Query: 41 ADVGVNAGLYAQELVANSARAIREEPKGSFNPVRVLEKAHSKTKAMGSSTVCIIALIDEA 100
A++ N ELVA ++ E GS+ VL S+ + S + I ++ E
Sbjct: 166 AELYYNDPTIFDELVAKNSNLRFSEDNGSY----VLRMPSSERIEINKSKIMIRSMEMEL 221
Query: 101 LNAINLGDSGFIVIRDGSVIFKSPVQQRGFNF 132
++ + G+ + I +KS +Q GFNF
Sbjct: 222 IHLVESGELDYFFI------YKSVAKQHGFNF 247
>PUPB_PSEPU (P38047) Ferric-pseudobactin BN7/BN8 receptor precursor
Length = 809
Score = 29.6 bits (65), Expect = 7.1
Identities = 33/105 (31%), Positives = 46/105 (43%), Gaps = 15/105 (14%)
Query: 108 DSGFIVIRDGSVIFKSPVQQRGFNFPYQLARSGTEGDLPSSGEVFTVPVAPGDIIVAGTD 167
D G ++ G+ + S RG N Y L S + G L S V APG GT
Sbjct: 97 DQGLAILLAGTGLEAS----RGANASYSLQASASTGALELSA-VSISGKAPGST-TEGT- 149
Query: 168 GLFDNMYNNDIVGVVVGATRARLGPQATAQKIAALARQRALDTKR 212
GL+ ++ +TR L P+ T Q + + RQR LD +R
Sbjct: 150 GLYTTYSSSS-------STRLNLTPRETPQSLTVMTRQR-LDDQR 186
>PDP2_RAT (O88484) [Pyruvate dehydrogenase [Lipoamide]]-phosphatase
2, mitochondrial precursor (EC 3.1.3.43) (PDP 2)
(Pyruvate dehydrogenase phosphatase, catalytic subunit
2) (PDPC 2)
Length = 530
Score = 29.6 bits (65), Expect = 7.1
Identities = 24/73 (32%), Positives = 41/73 (55%), Gaps = 4/73 (5%)
Query: 145 LPSSGEVFTVPVAPGD-IIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATA--QKIAA 201
L + EV + P D +V +DGL+D + N D+V +VVG +++G Q A Q+ A
Sbjct: 390 LTAKPEVTYHRLRPQDKFLVLASDGLWDMLDNEDVVRLVVGHL-SKVGHQKPALDQRPAN 448
Query: 202 LARQRALDTKRQS 214
L ++L +R++
Sbjct: 449 LGHMQSLLLQRKA 461
>MASZ_PSEPK (Q88QX8) Malate synthase G (EC 2.3.3.9)
Length = 725
Score = 29.6 bits (65), Expect = 7.1
Identities = 28/121 (23%), Positives = 49/121 (40%), Gaps = 10/121 (8%)
Query: 121 FKSPVQQRGFNFP----YQLARSGTEGDLPS-SGEVFTVPVAPGDIIVAGTDG----LFD 171
+K+ +Q+ G+ P +Q + ++ +G VPV + + L+D
Sbjct: 81 YKAFLQEIGYLLPQADDFQATTQNVDEEIAHMAGPQLVVPVMNARFALNAANARWGSLYD 140
Query: 172 NMYNNDIVGVVVGATRARLGPQATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGK 231
+Y D + GA + + + K+ A AR LD + GYR +GGK
Sbjct: 141 ALYGTDAISDEGGAEKGQGYNKVRGDKVIAFARA-FLDEAAPLAAGSHVDSTGYRIEGGK 199
Query: 232 L 232
L
Sbjct: 200 L 200
>UBA1_YEAST (P22515) Ubiquitin-activating enzyme E1 1
Length = 1024
Score = 29.3 bits (64), Expect = 9.3
Identities = 18/64 (28%), Positives = 32/64 (49%)
Query: 79 AHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFKSPVQQRGFNFPYQLAR 138
A+SK +GS + L + AL + G G+IV+ D I KS + ++ P + +
Sbjct: 433 ANSKVFLVGSGAIGCEMLKNWALLGLGSGSDGYIVVTDNDSIEKSNLNRQFLFRPKDVGK 492
Query: 139 SGTE 142
+ +E
Sbjct: 493 NKSE 496
>SYD_BACSU (O32038) Aspartyl-tRNA synthetase (EC 6.1.1.12)
(Aspartate--tRNA ligase) (AspRS)
Length = 592
Score = 29.3 bits (64), Expect = 9.3
Identities = 51/234 (21%), Positives = 90/234 (37%), Gaps = 23/234 (9%)
Query: 29 QAIGVADGVGGWADVGVNA-GLYAQELVANSARAIREEPKGSFNPVRVLEKAHSKTKAMG 87
+AI V G G ++ ++A G +A A ++ E G P+ ++K
Sbjct: 325 KAINVKGGAGDYSRKDIDALGAFAANYGAKGLAWVKVEADGVKGPIAKFFDEEKQSK--- 381
Query: 88 SSTVCIIALIDEALNAINL-GDSGFIVIRD--GSVIFK-----SPVQQRGFNFPYQ---- 135
+I +D A + L G F V+ G++ K + ++ FNF +
Sbjct: 382 -----LIEALDAAEGDLLLFGADQFEVVAASLGALRLKLGKERGLIDEKLFNFLWVIDWP 436
Query: 136 -LARSGTEGDLPSSGEVFTVPVAPG-DIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQ 193
L EG ++ FT+PV ++I + + Y+ + G +G R+ +
Sbjct: 437 LLEHDPEEGRFYAAHHPFTMPVREDLELIETAPEDMKAQAYDLVLNGYELGGGSIRIFEK 496
Query: 194 ATAQKIAALARQRALDTKRQSPFSAAALEYGYRFDGGKLDDLTVVVSYISNSVN 247
+K+ AL + Q F A EYG GG L +V ++ N
Sbjct: 497 DIQEKMFALLGFSPEEAAEQFGFLLEAFEYGAPPHGGIALGLDRLVMLLAGRTN 550
>PDP2_HUMAN (Q9P2J9) [Pyruvate dehydrogenase
[Lipoamide]]-phosphatase 2, mitochondrial precursor (EC
3.1.3.43) (PDP 2) (Pyruvate dehydrogenase phosphatase,
catalytic subunit 2) (PDPC 2)
Length = 529
Score = 29.3 bits (64), Expect = 9.3
Identities = 28/98 (28%), Positives = 46/98 (46%), Gaps = 8/98 (8%)
Query: 125 VQQRGFN------FPYQLARSGTEGDLPSSGEVFTVPVAPGD-IIVAGTDGLFDNMYNND 177
+ +RGFN + + T L + EV + P D +V +DGL+D + N D
Sbjct: 363 ILERGFNTEALNIYQFTPPHYYTPPYLTAEPEVTYHRLRPQDKFLVLASDGLWDMLSNED 422
Query: 178 IVGVVVG-ATRARLGPQATAQKIAALARQRALDTKRQS 214
+V +VVG A AQ+ A L ++L +R++
Sbjct: 423 VVRLVVGHLAEADWHKTDLAQRPANLGLMQSLLLQRKA 460
>HUPF_RHOCA (Q03005) Hydrogenase expression/formation protein hupF
Length = 106
Score = 29.3 bits (64), Expect = 9.3
Identities = 22/55 (40%), Positives = 26/55 (47%), Gaps = 5/55 (9%)
Query: 159 GDIIVAGTDGLFD-----NMYNNDIVGVVVGATRARLGPQATAQKIAALARQRAL 208
GD+I G GL D D V +GA R L P+A AQ AAL R+L
Sbjct: 17 GDVIEDGRPGLVDLSLVPEARPGDWVLAFLGAAREVLTPEAAAQISAALGGLRSL 71
>FAK1_CHICK (Q00944) Focal adhesion kinase 1 (EC 2.7.1.112) (FADK 1)
(PP125FAK)
Length = 1053
Score = 29.3 bits (64), Expect = 9.3
Identities = 26/139 (18%), Positives = 55/139 (38%), Gaps = 3/139 (2%)
Query: 79 AHSKTKAMGSSTVCIIALIDEALNAINLGDSGFIVIRDGSVIFKSPVQQRGFNFPYQLAR 138
A+ ++K + ++ A + + LGD G + S +K+ + +
Sbjct: 535 AYLESKRFVHRDIAARNVLVSATDCVKLGDFGLSRYMEDSTYYKA--SKGKLPIKWMAPE 592
Query: 139 SGTEGDLPSSGEVFTVPVAPGDIIVAGTDGLFDNMYNNDIVGVVVGATRARLGPQATAQK 198
S S+ +V+ V +I++ G F + NND++G + R + P
Sbjct: 593 SINFRRFTSASDVWMFGVCMWEILMHGVKP-FQGVKNNDVIGRIENGERLPMPPNCPPTL 651
Query: 199 IAALARQRALDTKRQSPFS 217
+ + + A D R+ F+
Sbjct: 652 YSLMTKCWAYDPSRRPRFT 670
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,485,663
Number of Sequences: 164201
Number of extensions: 1236717
Number of successful extensions: 2776
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 2765
Number of HSP's gapped (non-prelim): 21
length of query: 248
length of database: 59,974,054
effective HSP length: 107
effective length of query: 141
effective length of database: 42,404,547
effective search space: 5979041127
effective search space used: 5979041127
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC147178.4


