

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147014.3 + phase: 0
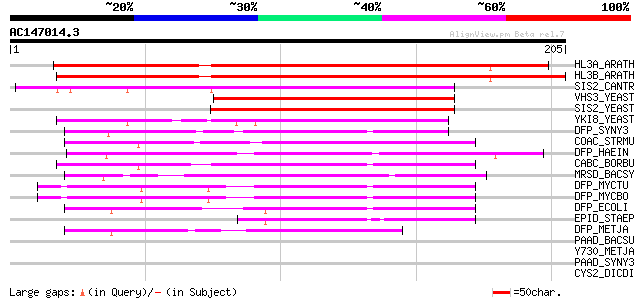
(205 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HL3A_ARATH (Q9SWE5) Phosphopantothenoylcysteine decarboxylase (E... 220 2e-57
HL3B_ARATH (P94063) Probable phosphopantothenoylcysteine decarbo... 213 2e-55
SIS2_CANTR (Q12600) SIS2 protein (Halotolerance protein HAL3) 110 3e-24
VHS3_YEAST (Q08438) Protein VHS3 (Viable in a HAL3 SIT4 backgrou... 101 1e-21
SIS2_YEAST (P36024) SIS2 protein (Halotolerance protein HAL3) 100 3e-21
YKI8_YEAST (P36076) Hypothetical protein YKL088w 96 4e-20
DFP_SYNY3 (P73881) Coenzyme A biosynthesis bifunctional protein ... 84 3e-16
COAC_STRMU (Q54433) Probable phosphopantothenoylcysteine decarbo... 73 4e-13
DFP_HAEIN (P44953) Coenzyme A biosynthesis bifunctional protein ... 72 9e-13
CABC_BORBU (O51752) Coenzyme A biosynthesis bifunctional protein... 69 1e-11
MRSD_BACSY (Q9RC23) Mersacidin decarboxylase (EC 4.1.1.-) (Mersa... 65 1e-10
DFP_MYCTU (P67733) Coenzyme A biosynthesis bifunctional protein ... 62 7e-10
DFP_MYCBO (P67734) Coenzyme A biosynthesis bifunctional protein ... 62 7e-10
DFP_ECOLI (P24285) Coenzyme A biosynthesis bifunctional protein ... 59 8e-09
EPID_STAEP (P30197) Epidermin decarboxylase (EC 4.1.1.-) (Epider... 57 2e-08
DFP_METJA (Q58323) Coenzyme A biosynthesis bifunctional protein ... 46 7e-05
PAAD_BACSU (P94404) Probable aromatic acid decarboxylase (EC 4.1... 42 0.001
Y730_METJA (Q58140) Hypothetical protein MJ0730 38 0.019
PAAD_SYNY3 (P72743) Probable aromatic acid decarboxylase (EC 4.1... 36 0.071
CYS2_DICDI (P04989) Cysteine proteinase 2 precursor (EC 3.4.22.-... 36 0.071
>HL3A_ARATH (Q9SWE5) Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (Halotolerance protein Hal3a) (AtHal3a)
(PPCDC) (AtCoaC)
Length = 209
Score = 220 bits (561), Expect = 2e-57
Identities = 108/187 (57%), Positives = 138/187 (73%), Gaps = 8/187 (4%)
Query: 17 PRKPRILLGCCGSVAAMKFGLVFNAFSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIY 76
PRKPR+LL GSVAA+KFG + + F+EWAEVRAVVT++SL FL T +Y
Sbjct: 17 PRKPRVLLAASGSVAAIKFGNLCHCFTEWAEVRAVVTKSSLHFLDKLSLPQEVT----LY 72
Query: 77 KDDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYE 136
D+ EW +W KIGD VLHIEL WAD++VIAPLSA+T KIAGGLCDNLLT I+RAWDY
Sbjct: 73 TDEDEWSSWNKIGDPVLHIELRRWADVLVIAPLSANTLGKIAGGLCDNLLTCIIRAWDYT 132
Query: 137 KPMFVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPPVQHM----PTNMREMADPSTIFST 192
KP+FVAP+M+ MW NPFTE++ +S++ELG+TLIPP++ MA+PS I+ST
Sbjct: 133 KPLFVAPAMNTLMWNNPFTERHLLSLDELGITLIPPIKKRLACGDYGNGAMAEPSLIYST 192
Query: 193 VKSFYDS 199
V+ F++S
Sbjct: 193 VRLFWES 199
>HL3B_ARATH (P94063) Probable phosphopantothenoylcysteine
decarboxylase (EC 4.1.1.36) (Halotolerance protein
Hal3b) (AtHal3b)
Length = 201
Score = 213 bits (542), Expect = 2e-55
Identities = 105/192 (54%), Positives = 137/192 (70%), Gaps = 8/192 (4%)
Query: 18 RKPRILLGCCGSVAAMKFGLVFNAFSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIYK 77
RKPRILL GSVA++KF + + FSEWAEV+AV +++SL F+ T +Y
Sbjct: 10 RKPRILLAASGSVASIKFSNLCHCFSEWAEVKAVASKSSLNFVDKPSLPQNVT----LYT 65
Query: 78 DDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEK 137
D+ EW +W KIGD VLHIEL WAD+M+IAPLSA+T AKIAGGLCDNLLT IVRAWDY K
Sbjct: 66 DEDEWSSWNKIGDPVLHIELRRWADVMIIAPLSANTLAKIAGGLCDNLLTCIVRAWDYSK 125
Query: 138 PMFVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPPVQHM----PTNMREMADPSTIFSTV 193
P+FVAP+M+ MW NPFTE++ + ++ELG+TLIPP++ MA+PS I+STV
Sbjct: 126 PLFVAPAMNTLMWNNPFTERHLVLLDELGITLIPPIKKKLACGDYGNGAMAEPSLIYSTV 185
Query: 194 KSFYDSNILKDK 205
+ F++S K +
Sbjct: 186 RLFWESQARKQR 197
>SIS2_CANTR (Q12600) SIS2 protein (Halotolerance protein HAL3)
Length = 531
Score = 110 bits (274), Expect = 3e-24
Identities = 61/177 (34%), Positives = 103/177 (57%), Gaps = 15/177 (8%)
Query: 3 ANSDSKSAIGHVNA---PRKPR------ILLGCCGSVAAMKFGLVFNAF-----SEWAEV 48
+NS++ + G N+ PR P+ +L+G CG+++ K L+ N S+ +
Sbjct: 242 SNSNTTTTKGEQNSNIDPRLPQDDGKFHVLIGVCGALSVGKVKLIVNKLLEIYTSDKISI 301
Query: 49 RAVVTETSLQFLVHEKAESLFTHRH-DIYKDDSEWKNWKKIGDSVLHIELANWADIMVIA 107
+ ++T++S FL+ E L + ++ D EW WK D VLHIEL WADI+++
Sbjct: 302 QVILTKSSENFLLPETLNVLENVKKVRVWTDIDEWTTWKTRLDPVLHIELRRWADILLVC 361
Query: 108 PLSAHTAAKIAGGLCDNLLTSIVRAWDYEKPMFVAPSMDGCMWRNPFTEQNFMSIEE 164
PL+A+T AKI+ G+CDNLLT+++RAW+ P+ +AP+MD + + T++ I +
Sbjct: 362 PLTANTLAKISLGICDNLLTNVIRAWNSSYPILLAPAMDSHSYSSSTTKRQLRLIAD 418
>VHS3_YEAST (Q08438) Protein VHS3 (Viable in a HAL3 SIT4 background
protein 3)
Length = 674
Score = 101 bits (252), Expect = 1e-21
Identities = 47/89 (52%), Positives = 65/89 (72%)
Query: 76 YKDDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDY 135
+ D EW W++ D VLHIEL WADI+V+APL+A+T AKIA GLCDNLLTS++RAW+
Sbjct: 441 WTDQDEWDVWRQRTDPVLHIELRRWADILVVAPLTANTLAKIALGLCDNLLTSVIRAWNP 500
Query: 136 EKPMFVAPSMDGCMWRNPFTEQNFMSIEE 164
P+F+APSM + + T+++F I+E
Sbjct: 501 TFPIFLAPSMGSGTFNSIMTKKHFRIIQE 529
>SIS2_YEAST (P36024) SIS2 protein (Halotolerance protein HAL3)
Length = 562
Score = 100 bits (248), Expect = 3e-21
Identities = 45/90 (50%), Positives = 64/90 (71%)
Query: 75 IYKDDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWD 134
++ D EW WK+ D VLHIEL WADI+V+APL+A+T +KIA GLCDNLLTS++RAW+
Sbjct: 359 LWTDQDEWDAWKQRTDPVLHIELRRWADILVVAPLTANTLSKIALGLCDNLLTSVIRAWN 418
Query: 135 YEKPMFVAPSMDGCMWRNPFTEQNFMSIEE 164
P+ +APSM + + T++ +I+E
Sbjct: 419 PSYPILLAPSMVSSTFNSMMTKKQLQTIKE 448
>YKI8_YEAST (P36076) Hypothetical protein YKL088w
Length = 571
Score = 96.3 bits (238), Expect = 4e-20
Identities = 58/157 (36%), Positives = 92/157 (57%), Gaps = 16/157 (10%)
Query: 18 RKPRILLGCCGSVAAMKFGLVFNAF-----SEWAEVRAVVTETSLQFLVHEKAESLFTHR 72
+K IL+G GSVA +K L+ + E ++ +VT+ + FL K + TH
Sbjct: 307 KKFHILIGATGSVATIKVPLIIDKLFKIYGPEKISIQLIVTKPAEHFL---KGLKMSTHV 363
Query: 73 HDIYKDDSEW------KNWKKIG-DSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNL 125
I++++ W KN + + +LH EL WADI +IAPLSA+T AK+A G+C+NL
Sbjct: 364 K-IWREEDAWVFDAVNKNDTSLSLNLILHHELRKWADIFLIAPLSANTLAKLANGICNNL 422
Query: 126 LTSIVRAWDYEKPMFVAPSMDGCMWRNPFTEQNFMSI 162
LTS++R W P+ +AP+M+ M+ NP T+++ S+
Sbjct: 423 LTSVMRDWSPLTPVLIAPAMNTFMYINPMTKKHLTSL 459
>DFP_SYNY3 (P73881) Coenzyme A biosynthesis bifunctional protein
coaBC (DNA/pantothenate metabolism flavoprotein)
[Includes: Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (PPCDC) (CoaC); Phosphopantothenate--cysteine
ligase (EC 6.3.2.5) (Phospho
Length = 402
Score = 83.6 bits (205), Expect = 3e-16
Identities = 53/143 (37%), Positives = 78/143 (54%), Gaps = 8/143 (5%)
Query: 21 RILLGCCGSVAAMKF-GLVFNAFSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIYKDD 79
RIL+G G +AA K +V F + AEVR ++T + +F+ +L RH Y D
Sbjct: 6 RILIGVGGGIAAYKICEVVSQLFQQGAEVRVILTAEAEKFVTPLTFTTLA--RHPAYGDA 63
Query: 80 SEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEKPM 139
W+ I LHI+L WADI +IAPL+AHT AK+ G D+LL++ V A P+
Sbjct: 64 DFWQ---PIHHRPLHIDLGEWADIFLIAPLTAHTLAKLGHGFADDLLSNTVLA--SSCPI 118
Query: 140 FVAPSMDGCMWRNPFTEQNFMSI 162
+AP+M+ MW ++N +
Sbjct: 119 LLAPAMNTDMWEQEAVQRNLQQL 141
>COAC_STRMU (Q54433) Probable phosphopantothenoylcysteine
decarboxylase (EC 4.1.1.36) (PPCDC)
Length = 179
Score = 73.2 bits (178), Expect = 4e-13
Identities = 48/153 (31%), Positives = 81/153 (52%), Gaps = 7/153 (4%)
Query: 21 RILLGCCGSVAAMKFGLVFNAFSEWA-EVRAVVTETSLQFLVHEKAESLFTHRHDIYKDD 79
+ILL GS+AA K + + ++ V ++T + QF+ + L ++ +Y +
Sbjct: 4 KILLAVSGSIAAYKAADLSHQLTKLGYHVNVLMTNAAKQFIPPLTLQVL--SKNPVYSNV 61
Query: 80 SEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEKPM 139
+ + + I HI LA AD+ ++AP SA+T A +A G DN++TS+ A E P
Sbjct: 62 MKEDDPQVIN----HIALAKQADLFLLAPASANTLAHLAHGFADNIVTSVALALPLEVPK 117
Query: 140 FVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPP 172
F AP+M+ M+ NP T+ N +++ G I P
Sbjct: 118 FFAPAMNTKMYENPITQSNIALLKKFGYKEIQP 150
>DFP_HAEIN (P44953) Coenzyme A biosynthesis bifunctional protein
coaBC (DNA/pantothenate metabolism flavoprotein)
[Includes: Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (PPCDC) (CoaC); Phosphopantothenate--cysteine
ligase (EC 6.3.2.5) (Phospho
Length = 400
Score = 72.0 bits (175), Expect = 9e-13
Identities = 52/181 (28%), Positives = 93/181 (50%), Gaps = 13/181 (7%)
Query: 22 ILLGCCGSVAAMK-FGLVFNAFSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIYKDDS 80
I++G G +AA K L+ AEVR V+T + +F+ +++ + D
Sbjct: 8 IVVGITGGIAAYKTIELIRLLRKAEAEVRVVLTPAAAEFVTPLTLQAISGNAVSQSLLDP 67
Query: 81 EWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEKPMF 140
+ + ++ HIELA WAD ++IAP SA A++ G+ ++LL++I A + P+F
Sbjct: 68 QAEL------AMGHIELAKWADAIIIAPASADFIARLTIGMANDLLSTICLATN--APIF 119
Query: 141 VAPSMDGCMWRNPFTEQNFMSIEELGVTLIPPVQHMPT----NMREMADPSTIFSTVKSF 196
+AP+M+ M+ T+QN +++ G+ LI P M++P IF+ + F
Sbjct: 120 LAPAMNQQMYHQSITQQNLTTLQTRGIELIGPNSGFQACGDMGKGRMSEPEEIFTALSDF 179
Query: 197 Y 197
+
Sbjct: 180 F 180
>CABC_BORBU (O51752) Coenzyme A biosynthesis bifunctional protein
coaBC (DNA/pantothenate metabolism flavoprotein)
[Includes: Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (PPCDC) (CoaC); Phosphopantothenate--cysteine
ligase (EC 6.3.2.5) (Phosph
Length = 390
Score = 68.6 bits (166), Expect = 1e-11
Identities = 40/156 (25%), Positives = 80/156 (50%), Gaps = 10/156 (6%)
Query: 18 RKPRILLGCCGSVAAMKFGLVFNAFSEWA-EVRAVVTETSLQFLVHEKAESLFTHRHDIY 76
+ IL+G CG +A+ K + ++ + +V+ ++T+ + +F+ E+ I
Sbjct: 3 KNKHILIGICGGIASYKSVYIVSSLVKLGYKVKVIMTQNATKFITPLTLET-------IS 55
Query: 77 KDDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYE 136
K+ W + V HI++A WA ++++ P + +T +KIA G+ D+ LT+I+ A
Sbjct: 56 KNKIITNLWDLDHNEVEHIKIAKWAHLILVIPATYNTISKIASGIADDALTTIISA--ST 113
Query: 137 KPMFVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPP 172
P + A +M+ M+ NP ++N ++ I P
Sbjct: 114 APTYFAIAMNNIMYSNPILKENIKKLKTYNYKFIEP 149
>MRSD_BACSY (Q9RC23) Mersacidin decarboxylase (EC 4.1.1.-)
(Mersacidin modifying enzyme mrsD)
Length = 194
Score = 65.1 bits (157), Expect = 1e-10
Identities = 45/159 (28%), Positives = 76/159 (47%), Gaps = 16/159 (10%)
Query: 21 RILLGCCGSVAAM---KFGLVFNAFSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIYK 77
++L+G CGS++++ + L F +F + E+R V+T+T AE L Y
Sbjct: 10 KLLIGICGSISSVGISSYLLYFKSF--FKEIRVVMTKT---------AEDLIPAHTVSYF 58
Query: 78 DDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEK 137
D + + G H+E+ WADI I P +A+ + A G+ NL+ + V A +
Sbjct: 59 CDHVYSEHGENGKRHSHVEIGRWADIYCIIPATANILGQTANGVAMNLVATTVLAHPHNT 118
Query: 138 PMFVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPPVQHM 176
F P+M+ MW +N + + G +I PV+ M
Sbjct: 119 IFF--PNMNDLMWNKTVVSRNIEQLRKDGHIVIEPVEIM 155
>DFP_MYCTU (P67733) Coenzyme A biosynthesis bifunctional protein
coaBC (DNA/pantothenate metabolism flavoprotein)
[Includes: Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (PPCDC) (CoaC); Phosphopantothenate--cysteine
ligase (EC 6.3.2.5) (Phospho
Length = 418
Score = 62.4 bits (150), Expect = 7e-10
Identities = 46/165 (27%), Positives = 80/165 (47%), Gaps = 17/165 (10%)
Query: 11 IGHVNAPRKPRILLGCCGSVAAMKFGLVFNAFSEWAE-VRAVVTETSLQFLVHEKAESLF 69
+ H P++ +++G G +AA K V +E + VR + TE++L+F+ E+L
Sbjct: 2 VDHKRIPKQ--VIVGVSGGIAAYKACTVVRQLTEASHRVRVIPTESALRFVGAATFEALS 59
Query: 70 THR--HDIYKDDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLT 127
D++ D +V H+ L AD++V+AP +A A+ A G D+LLT
Sbjct: 60 GEPVCTDVFADVP----------AVPHVHLGQQADLVVVAPATADLLARAAAGRADDLLT 109
Query: 128 SIVRAWDYEKPMFVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPP 172
+ + P+ AP+M MW +P T N ++ G ++ P
Sbjct: 110 ATLLT--ARCPVLFAPAMHTEMWLHPATVDNVATLRRRGAVVLEP 152
>DFP_MYCBO (P67734) Coenzyme A biosynthesis bifunctional protein
coaBC (DNA/pantothenate metabolism flavoprotein)
[Includes: Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (PPCDC) (CoaC); Phosphopantothenate--cysteine
ligase (EC 6.3.2.5) (Phospho
Length = 418
Score = 62.4 bits (150), Expect = 7e-10
Identities = 46/165 (27%), Positives = 80/165 (47%), Gaps = 17/165 (10%)
Query: 11 IGHVNAPRKPRILLGCCGSVAAMKFGLVFNAFSEWAE-VRAVVTETSLQFLVHEKAESLF 69
+ H P++ +++G G +AA K V +E + VR + TE++L+F+ E+L
Sbjct: 2 VDHKRIPKQ--VIVGVSGGIAAYKACTVVRQLTEASHRVRVIPTESALRFVGAATFEALS 59
Query: 70 THR--HDIYKDDSEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLT 127
D++ D +V H+ L AD++V+AP +A A+ A G D+LLT
Sbjct: 60 GEPVCTDVFADVP----------AVPHVHLGQQADLVVVAPATADLLARAAAGRADDLLT 109
Query: 128 SIVRAWDYEKPMFVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPP 172
+ + P+ AP+M MW +P T N ++ G ++ P
Sbjct: 110 ATLLT--ARCPVLFAPAMHTEMWLHPATVDNVATLRRRGAVVLEP 152
>DFP_ECOLI (P24285) Coenzyme A biosynthesis bifunctional protein
coaBC (DNA/pantothenate metabolism flavoprotein)
[Includes: Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (PPCDC) (CoaC); Phosphopantothenate--cysteine
ligase (EC 6.3.2.5) (Phospho
Length = 405
Score = 58.9 bits (141), Expect = 8e-09
Identities = 44/162 (27%), Positives = 78/162 (47%), Gaps = 27/162 (16%)
Query: 21 RILLGCCGSVAAMKFG-LVFNAFSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIYKDD 79
+I+LG G +AA K LV A+VR +TE + F+ +++ +
Sbjct: 6 KIVLGVSGGIAAYKTPELVRRLRDRGADVRVAMTEAAKAFITPLSLQAVSGY-------- 57
Query: 80 SEWKNWKKIGDSVL---------HIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIV 130
+ DS+L HIEL WAD++++AP +A A++A G+ ++L+++I
Sbjct: 58 -------PVSDSLLDPAAEAAMGHIELGKWADLVILAPATADLIARVAAGMANDLVSTIC 110
Query: 131 RAWDYEKPMFVAPSMDGCMWRNPFTEQNFMSIEELGVTLIPP 172
A P+ V P+M+ M+R T+ N + G+ + P
Sbjct: 111 LA--TPAPVAVLPAMNQQMYRAAATQHNLEVLASRGLLIWGP 150
>EPID_STAEP (P30197) Epidermin decarboxylase (EC 4.1.1.-) (Epidermin
modifying enzyme epiD)
Length = 181
Score = 57.4 bits (137), Expect = 2e-08
Identities = 32/89 (35%), Positives = 54/89 (59%), Gaps = 3/89 (3%)
Query: 85 WKKIGDSVL-HIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEKPMFVAP 143
+ +I D +L HI + + +++ P SA+T KIA G+CDNLLT++ Y+K +F+ P
Sbjct: 57 YDEIKDPLLNHINIVENHEYILVLPASANTINKIANGICDNLLTTVCLT-GYQK-LFIFP 114
Query: 144 SMDGCMWRNPFTEQNFMSIEELGVTLIPP 172
+M+ MW NPF ++N ++ V + P
Sbjct: 115 NMNIRMWGNPFLQKNIDLLKNNDVKVYSP 143
>DFP_METJA (Q58323) Coenzyme A biosynthesis bifunctional protein
coaBC (DNA/pantothenate metabolism flavoprotein)
[Includes: Phosphopantothenoylcysteine decarboxylase (EC
4.1.1.36) (PPCDC) (CoaC); Phosphopantothenate--cysteine
ligase (EC 6.3.2.5) (Phospho
Length = 403
Score = 45.8 bits (107), Expect = 7e-05
Identities = 33/126 (26%), Positives = 64/126 (50%), Gaps = 12/126 (9%)
Query: 21 RILLGCCGSVAAMKFG-LVFNAFSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIYKDD 79
+IL+ S+AA++ L+ AEV ++TE + + + E + F +++Y++
Sbjct: 24 KILVAVTSSIAAIETPKLMRELIRHGAEVYCIITEETKKIIGKEALK--FGCGNEVYEE- 80
Query: 80 SEWKNWKKIGDSVLHIELANWADIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEKPM 139
I + HI L N D ++I P +A+ +KI G+ DN++ + + KP+
Sbjct: 81 --------ITGDIEHILLYNECDCLLIYPATANIISKINLGIADNIVNTTALMFFGNKPI 132
Query: 140 FVAPSM 145
F+ P+M
Sbjct: 133 FIVPAM 138
>PAAD_BACSU (P94404) Probable aromatic acid decarboxylase (EC
4.1.1.-)
Length = 204
Score = 42.0 bits (97), Expect = 0.001
Identities = 27/98 (27%), Positives = 49/98 (49%), Gaps = 6/98 (6%)
Query: 102 DIMVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYEKPMFVAPSMDGCMWRNPFTEQNFMS 161
D M++AP S + A I G+ DNLLT E+ V + + + N +N ++
Sbjct: 90 DGMIVAPCSMKSLASIRTGMADNLLTRAADVMLKERKKLVLLTRETPL--NQIHLENMLA 147
Query: 162 IEELGVTLIPPV---QHMPTNMREMADPSTIFSTVKSF 196
+ ++G ++PP+ + P ++ EM D +F T+ F
Sbjct: 148 LTKMGTIILPPMPAFYNRPRSLEEMVD-HIVFRTLDQF 184
>Y730_METJA (Q58140) Hypothetical protein MJ0730
Length = 186
Score = 37.7 bits (86), Expect = 0.019
Identities = 17/43 (39%), Positives = 30/43 (69%), Gaps = 1/43 (2%)
Query: 102 DIMVIAPLSAHTAAKIAGGLCDNLLT-SIVRAWDYEKPMFVAP 143
D+ ++AP +A+T AKIA G+ D L+T S+ +A + P+++ P
Sbjct: 82 DLFLVAPATANTTAKIAYGIADTLITNSVAQAMKAKVPVYIFP 124
>PAAD_SYNY3 (P72743) Probable aromatic acid decarboxylase (EC
4.1.1.-)
Length = 206
Score = 35.8 bits (81), Expect = 0.071
Identities = 28/92 (30%), Positives = 44/92 (47%), Gaps = 19/92 (20%)
Query: 104 MVIAPLSAHTAAKIAGGLCDNLLTSIVRAWDYE----KPMFVAPSMDGCMWRNPFT---E 156
MV+ P S T AK+A G+ +LL RA D + KP+ V P P +
Sbjct: 97 MVVLPCSMSTVAKLAVGMSSDLLE---RAADVQIKEGKPLVVVPR------ETPLSLIHL 147
Query: 157 QNFMSIEELGVTLIPPV---QHMPTNMREMAD 185
+N S+ E GV ++P + H P ++ ++ D
Sbjct: 148 RNLTSLAEAGVRIVPAIPAWYHQPQSVEDLVD 179
>CYS2_DICDI (P04989) Cysteine proteinase 2 precursor (EC 3.4.22.-)
(Prestalk cathepsin)
Length = 376
Score = 35.8 bits (81), Expect = 0.071
Identities = 23/63 (36%), Positives = 38/63 (59%), Gaps = 3/63 (4%)
Query: 42 FSEWAEVRAVVTETSLQFLVHEKAESLFTHRHDIYKDDSEW-KNWKKIGDSVLHIELANW 100
FSE ++ R TE +L+F + + S F++R+ I+K + ++ NW GDS + L N+
Sbjct: 27 FSE-SQYRTAFTEWTLKFN-RQYSSSEFSNRYSIFKSNMDYVDNWNSKGDSQTVLGLNNF 84
Query: 101 ADI 103
ADI
Sbjct: 85 ADI 87
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.133 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,885,168
Number of Sequences: 164201
Number of extensions: 884859
Number of successful extensions: 1831
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1788
Number of HSP's gapped (non-prelim): 37
length of query: 205
length of database: 59,974,054
effective HSP length: 105
effective length of query: 100
effective length of database: 42,732,949
effective search space: 4273294900
effective search space used: 4273294900
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC147014.3


