

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147013.12 - phase: 0
(687 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
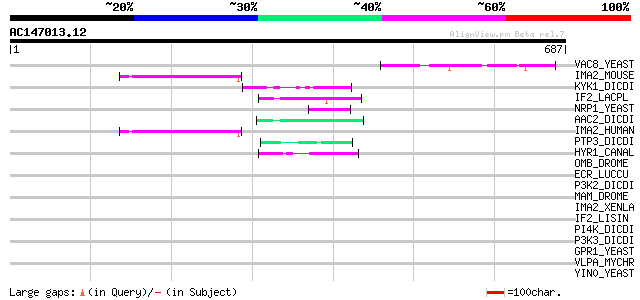
Score E
Sequences producing significant alignments: (bits) Value
VAC8_YEAST (P39968) Vacuolar protein 8 61 1e-08
IMA2_MOUSE (P52293) Importin alpha-2 subunit (Karyopherin alpha-... 51 9e-06
KYK1_DICDI (P18160) Non-receptor tyrosine kinase spore lysis A (... 49 3e-05
IF2_LACPL (Q88VK7) Translation initiation factor IF-2 49 3e-05
NRP1_YEAST (P32770) Asparagine-rich protein (ARP protein) 48 8e-05
AAC2_DICDI (P14196) AAC-rich mRNA clone AAC11 protein (Fragment) 48 8e-05
IMA2_HUMAN (P52292) Importin alpha-2 subunit (Karyopherin alpha-... 47 2e-04
PTP3_DICDI (P54637) Protein-tyrosine phosphatase 3 (EC 3.1.3.48)... 47 2e-04
HYR1_CANAL (P46591) Hyphally regulated protein precursor 47 2e-04
OMB_DROME (Q24432) Optomotor-blind protein (Lethal(1)optomotor-b... 44 0.001
ECR_LUCCU (O18531) Ecdysone receptor (Ecdysteroid receptor) (20-... 44 0.001
P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.13... 44 0.002
MAM_DROME (P21519) Neurogenic protein mastermind 44 0.002
IMA2_XENLA (P52171) Importin alpha-2 subunit (Karyopherin alpha-... 44 0.002
IF2_LISIN (Q92C29) Translation initiation factor IF-2 43 0.003
PI4K_DICDI (P54677) Phosphatidylinositol 4-kinase (EC 2.7.1.67) ... 42 0.004
P3K3_DICDI (P54675) Phosphatidylinositol 3-kinase 3 (EC 2.7.1.13... 42 0.004
GPR1_YEAST (Q12361) G protein-coupled receptor GPR1 42 0.004
VLPA_MYCHR (P29228) Variant surface antigen A precursor (VlpA pr... 42 0.005
YIN0_YEAST (P40467) Putative 108.8 kDa transcriptional regulator... 42 0.007
>VAC8_YEAST (P39968) Vacuolar protein 8
Length = 578
Score = 60.8 bits (146), Expect = 1e-08
Identities = 57/229 (24%), Positives = 102/229 (43%), Gaps = 29/229 (12%)
Query: 460 ITESRALLCFSVLLEKGPEAVQYNSAMALMEITAVAEKDAELRKSAFKPNSPACKAVVDQ 519
I S AL+ + L + VQ N+ AL+ +T E EL + P
Sbjct: 163 IATSGALIPLTKLAKSKHIRVQRNATGALLNMTHSEENRKELVNAGAVP----------V 212
Query: 520 VLKIIEKADSDLLIPCVKAIGNLA------RTFKATETRMIGPLVKLLDEREAEVSREAS 573
++ ++ D D+ C A+ N+A + TE R++ LV L+D + V +A+
Sbjct: 213 LVSLLSSTDPDVQYYCTTALSNIAVDEANRKKLAQTEPRLVSKLVSLMDSPSSRVKCQAT 272
Query: 574 IALRKFAGSENYLHVDHSNAIISAGGAKHLIQLVYFGEQMVQIPALVLLSYIALHVPDSE 633
+ALR A +Y I+ AGG HL++L+ + + ++ + I++H P +E
Sbjct: 273 LALRNLASDTSY-----QLEIVRAGGLPHLVKLIQSDSIPLVLASVACIRNISIH-PLNE 326
Query: 634 ELA-----LAEVLGVLEWASKQSFMQHD-ETLEELLQEA-KSRLELYQS 675
L L ++ +L++ + H TL L + K+R E ++S
Sbjct: 327 GLIVDAGFLKPLVRLLDYKDSEEIQCHAVSTLRNLAASSEKNRKEFFES 375
>IMA2_MOUSE (P52293) Importin alpha-2 subunit (Karyopherin alpha-2
subunit) (SRP1-alpha) (RAG cohort protein 1) (Pendulin)
(Pore targeting complex 58 kDa subunit) (PTAC58)
(Importin alpha P1)
Length = 529
Score = 51.2 bits (121), Expect = 9e-06
Identities = 42/157 (26%), Positives = 74/157 (46%), Gaps = 9/157 (5%)
Query: 136 PPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLK 195
PP+ A E IL + + +L EV +D+ ++ L G + ++++++G V L+K
Sbjct: 244 PPLDAVEQILPTL---VRLLHHNDPEVLADSCWAISYLTDGPNERIEMVVKKGVVPQLVK 300
Query: 196 LIKEGKADGQENAARAIG--LLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAV 253
L+ + A RAIG + G D E + +I G +VF +L +Q W +
Sbjct: 301 LLGATELPIVTPALRAIGNIVTGTD-EQTQKVIDAGALAVFPSLLTNPKTNIQKEATWTM 359
Query: 254 SELAANYPKCQELFAQHNIIRLLVGHLA---FETVEE 287
S + A + H ++ LVG L+ F+T +E
Sbjct: 360 SNITAGRQDQIQQVVNHGLVPFLVGVLSKADFKTQKE 396
Score = 32.0 bits (71), Expect = 5.6
Identities = 28/151 (18%), Positives = 60/151 (39%), Gaps = 2/151 (1%)
Query: 153 AMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARAI 212
++L ++ +A ++ ++ G + ++ G V L+ ++ + Q+ AA AI
Sbjct: 342 SLLTNPKTNIQKEATWTMSNITAGRQDQIQQVVNHGLVPFLVGVLSKADFKTQKEAAWAI 401
Query: 213 G--LLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVSELAANYPKCQELFAQH 270
G E + +++H G+ +L K+ V+ A+S + K E
Sbjct: 402 TNYTSGGTVEQIVYLVHCGIIEPLMNLLSAKDTKIIQVILDAISNIFQAAEKLGETEKLS 461
Query: 271 NIIRLLVGHLAFETVEEHSKYAIVSMKANSI 301
+I G E ++ H ++ N I
Sbjct: 462 IMIEECGGLDKIEALQRHENESVYKASLNLI 492
>KYK1_DICDI (P18160) Non-receptor tyrosine kinase spore lysis A (EC
2.7.1.112) (Tyrosine-protein kinase 1)
Length = 1584
Score = 49.3 bits (116), Expect = 3e-05
Identities = 35/135 (25%), Positives = 57/135 (41%), Gaps = 32/135 (23%)
Query: 289 SKYAIVSMKANSIHAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGE 348
S+ ++ NS V +NNNNS++N N N + ++G G
Sbjct: 385 SQVTLMLKNVNSTSILVPNGNNNNNSNNNNN------NNNNNIIGNG------------- 425
Query: 349 RPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQG 408
++ T+T S P+ N N++I +N+N N N N N N N+
Sbjct: 426 ------KITTTT----TTSTSPSS---INNNEDISSNNNNNNNNNNNNNNNNNNNNNNNN 472
Query: 409 NHNNHQRNYSHSGIN 423
N+NN+ N S++ N
Sbjct: 473 NNNNNNSNSSNTNNN 487
Score = 34.7 bits (78), Expect = 0.86
Identities = 27/124 (21%), Positives = 49/124 (38%), Gaps = 13/124 (10%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
+NNNN+++N N N + N + +++ N + S + +
Sbjct: 461 NNNNNNNNNNNNNNNNNNSNSSNTNNNNINNTTNNNNSNSNNNNNNNNSNSNSNSNNNNI 520
Query: 369 QPNEGNEANQNQNI-------LANSNTPNGNGLGNGNGNGNDGGKQGNHNNH-----QRN 416
N N N N NI + +++ + LG N +GN+ G+ N+ QR+
Sbjct: 521 NNNNNNN-NNNNNIYLTKKPSIGSTDESSTGSLGGNNSSGNNNSSSGSIGNNSSIIKQRS 579
Query: 417 YSHS 420
HS
Sbjct: 580 PPHS 583
>IF2_LACPL (Q88VK7) Translation initiation factor IF-2
Length = 858
Score = 49.3 bits (116), Expect = 3e-05
Identities = 34/132 (25%), Positives = 55/132 (40%), Gaps = 11/132 (8%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
SNN N S+N + N GG+ R +++ R N +R TS ++
Sbjct: 125 SNNGNRSANASRSNSARNN------GGSNTNRSNNNNNNNRSNNNNRSNTSNNRTNSTGN 178
Query: 369 QPNEGNEANQNQNILANSNTPN---GNGLGNG--NGNGNDGGKQGNHNNHQRNYSHSGIN 423
+ N G + N N+N N NG N NG+G GG ++NN Y N
Sbjct: 179 RTNNGGQNRNNNRTTTNNNNNNSNRSNGSNNSSRNGSGRFGGSLNSNNNGGGRYRGGNNN 238
Query: 424 MKGRESEDAETK 435
+ R + + +++
Sbjct: 239 NRRRNNRNNKSR 250
Score = 40.0 bits (92), Expect = 0.020
Identities = 29/127 (22%), Positives = 48/127 (36%), Gaps = 7/127 (5%)
Query: 311 NNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASKQP 370
NNN ++N N + + GGN+ ++ G R N R S A +
Sbjct: 96 NNNRNNNQNNSQSRNSNGNRSNNGGNR----TNSNNGNRSANASR---SNSARNNGGSNT 148
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESE 430
N N N N N+ + N N GN + G Q +NN +++ + + S
Sbjct: 149 NRSNNNNNNNRSNNNNRSNTSNNRTNSTGNRTNNGGQNRNNNRTTTNNNNNNSNRSNGSN 208
Query: 431 DAETKAS 437
++ S
Sbjct: 209 NSSRNGS 215
Score = 32.3 bits (72), Expect = 4.3
Identities = 20/58 (34%), Positives = 28/58 (47%), Gaps = 6/58 (10%)
Query: 367 SKQPNEGNEANQNQNILANSNTPNGNGLGNGNGN-GNDGGKQGNHNNHQRNYSHSGIN 423
S+ G+ N N+N N N + N NGN N+GG + N NN R+ + S N
Sbjct: 86 SRHQQSGDYLNNNRN-----NNQNNSQSRNSNGNRSNNGGNRTNSNNGNRSANASRSN 138
>NRP1_YEAST (P32770) Asparagine-rich protein (ARP protein)
Length = 719
Score = 48.1 bits (113), Expect = 8e-05
Identities = 22/51 (43%), Positives = 26/51 (50%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSG 421
N N N N+ N N N GNGNGNGN+ NHNN+ N H+G
Sbjct: 492 NINNNNNNINNMTNNRYNINNNINGNGNGNGNNSNNNNNHNNNHNNNHHNG 542
Score = 43.1 bits (100), Expect = 0.002
Identities = 20/48 (41%), Positives = 24/48 (49%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYS 418
N N N N +N NGNG GNGN + N+ NHNN+ N S
Sbjct: 496 NNNNINNMTNNRYNINNNINGNGNGNGNNSNNNNNHNNNHNNNHHNGS 543
Score = 40.4 bits (93), Expect = 0.016
Identities = 39/180 (21%), Positives = 65/180 (35%), Gaps = 28/180 (15%)
Query: 253 VSELAANYPKCQELF---AQHNIIRLLVGHLAFETVEEHSKYAIVSMKANSIHAAVVMA- 308
+S AAN+P F A + L F + S+ ++M ++ + +++A
Sbjct: 416 ISNTAANHPYGAPEFNMIANNTPAALTYNRAHFPAITPLSRQNSLNMAPSNSGSPIIIAD 475
Query: 309 --SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAA 366
S NNN + N N + + N ++++ G
Sbjct: 476 HFSGNNNIAPNYRYNNNINNNNNNINNMTNNRYNINNNINGN------------------ 517
Query: 367 SKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKG 426
GN N N N N+N N + G+ N N N N+N + N +S I M G
Sbjct: 518 ----GNGNGNNSNNNNNHNNNHNNNHHNGSINSNSNTNNNNNNNNGNNSNNCNSNIGMGG 573
Score = 32.3 bits (72), Expect = 4.3
Identities = 39/184 (21%), Positives = 68/184 (36%), Gaps = 22/184 (11%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
+NNNN+++N N + G+ G N R H + + +
Sbjct: 550 TNNNNNNNNGNNSNNCNSNIGMGGCGSNMPFRAGDWKCSTC--TYHNFAKNVVCLRCGGP 607
Query: 369 QPNEGNEANQNQNILAN-----SNTPNGNGLG-NGNGNGNDGGKQGNHNNHQRNYSHSGI 422
+ G+ + N I ++ S TP+ N + N NG N G GN N
Sbjct: 608 KSISGDASETNHYIDSSTFGPASRTPSNNNISVNTNGGSNAGRTDGNDN----------- 656
Query: 423 NMKGRESEDAETKASMKEMAARALWHLAKGNVAICRSITESRALLCFSVLLEKGPEAVQY 482
KGR+ E + MA +++ GN + +A + FS + + +
Sbjct: 657 --KGRDISLMEFMSPPLSMATKSMKE-GDGNGSSFNEFKSDKANVNFSNVGDNSAFGNGF 713
Query: 483 NSAM 486
NS++
Sbjct: 714 NSSI 717
>AAC2_DICDI (P14196) AAC-rich mRNA clone AAC11 protein (Fragment)
Length = 448
Score = 48.1 bits (113), Expect = 8e-05
Identities = 32/132 (24%), Positives = 52/132 (39%), Gaps = 8/132 (6%)
Query: 306 VMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHA 365
++ SNNNN+++N N N N + +++ N + T+ +
Sbjct: 266 ILNSNNNNNNNNNNSNINNIN--------NNNNNNSNNNNNSSNNNNNNNNSTNNNTNNN 317
Query: 366 ASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMK 425
+ N N N N N N+ N N N N N N+ G N+NN+ N +H N
Sbjct: 318 NNNTNNNTNNNNNNINNNNNNTNNNNNNANNQNTNNNNMGNNSNNNNNPNNNNHQNNNNN 377
Query: 426 GRESEDAETKAS 437
+ T A+
Sbjct: 378 NTSNNSNTTTAT 389
Score = 39.7 bits (91), Expect = 0.027
Identities = 32/121 (26%), Positives = 48/121 (39%), Gaps = 17/121 (14%)
Query: 308 ASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAAS 367
++NNN +++N N T N + + N +++ + N + S +
Sbjct: 309 STNNNTNNNNNNTNNNTNNNNNNINNNNNNTNNNNNNANNQNTNNNNMGNNSN-----NN 363
Query: 368 KQPNEGNEANQNQNILA-NSNT-------PNGNGLGNGNGN----GNDGGKQGNHNNHQR 415
PN N N N N + NSNT P GN L N N GN G G H++
Sbjct: 364 NNPNNNNHQNNNNNNTSNNSNTTTATTTAPGGNNLTNSLNNAGNLGNLGRVSGLHSSDPN 423
Query: 416 N 416
N
Sbjct: 424 N 424
Score = 37.7 bits (86), Expect = 0.10
Identities = 32/138 (23%), Positives = 50/138 (36%), Gaps = 8/138 (5%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
SNNNN+SSN N + N + +++ N + + A + +
Sbjct: 293 SNNNNNSSNNNNNNNNSTNNNTNNNNNNTNNNTNNNNNNINNNNNNTNNNNNNANNQNTN 352
Query: 369 QPNEGNEANQNQNILANSNTPNGNGLGNGNGN-------GNDGGKQGNHNNHQRNYSHSG 421
N GN +N N N N++ N N + N N G N N+ N + G
Sbjct: 353 NNNMGNNSNNNNNPNNNNHQNNNNNNTSNNSNTTTATTTAPGGNNLTNSLNNAGNLGNLG 412
Query: 422 INMKGRESEDAETKASMK 439
+ G S D + K
Sbjct: 413 -RVSGLHSSDPNNPNAQK 429
Score = 36.6 bits (83), Expect = 0.23
Identities = 28/110 (25%), Positives = 42/110 (37%), Gaps = 16/110 (14%)
Query: 309 SNNNNSSSNLNPKKGTENEDG--VVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAA 366
S+ NN+S + + GT + G G + +H+ LG N
Sbjct: 223 SDYNNTSFSDSNTDGTPKKRGRPPKAKGESPSASPTHNTLGNGILN-------------- 268
Query: 367 SKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRN 416
S N N N N N + N+N N N N + N N+ N+N + N
Sbjct: 269 SNNNNNNNNNNSNINNINNNNNNNSNNNNNSSNNNNNNNNSTNNNTNNNN 318
Score = 36.6 bits (83), Expect = 0.23
Identities = 27/105 (25%), Positives = 45/105 (42%), Gaps = 1/105 (0%)
Query: 331 VVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPN 390
++GGGN +G S + G+ + + ++AI S P N N N N +N N N
Sbjct: 73 MMGGGN-NGNNSGNNNGQPQMHGTNLNGLSLAIQNQSSLPQPINNNNNNNNNNSNINNNN 131
Query: 391 GNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETK 435
N N N N ++ G + N + + R++ +E K
Sbjct: 132 NNSNNNNNNNNSNLGINSSPTQSSANSADKRSRGRPRKNPPSEPK 176
Score = 36.6 bits (83), Expect = 0.23
Identities = 38/171 (22%), Positives = 57/171 (33%), Gaps = 48/171 (28%)
Query: 309 SNNNNSSSNLN--------------------PKKGTENEDGVVVGGGNKHGRVSHHPLGE 348
+NNNN++SNL P+K +E G K GR
Sbjct: 136 NNNNNNNSNLGINSSPTQSSANSADKRSRGRPRKNPPSEPKDTSGPKRKRGRPPKMDEEG 195
Query: 349 RPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNT-------------------- 388
P+ + K+P + NE++ N ++SNT
Sbjct: 196 NPQPKPVPQPGSNKKRGRPKKPKDENESDYNNTSFSDSNTDGTPKKRGRPPKAKGESPSA 255
Query: 389 -PNGNGLGNG-------NGNGNDGGKQGNHNNHQRNYSHSGINMKGRESED 431
P N LGNG N N N+ N NN+ N S++ N + +
Sbjct: 256 SPTHNTLGNGILNSNNNNNNNNNNSNINNINNNNNNNSNNNNNSSNNNNNN 306
>IMA2_HUMAN (P52292) Importin alpha-2 subunit (Karyopherin alpha-2
subunit) (SRP1-alpha) (RAG cohort protein 1)
Length = 529
Score = 47.0 bits (110), Expect = 2e-04
Identities = 42/157 (26%), Positives = 70/157 (43%), Gaps = 9/157 (5%)
Query: 136 PPIAANEPILCFIWEQIAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLK 195
PPI A E IL + + +L EV +D ++ L G + ++++ G V L+K
Sbjct: 244 PPIDAVEQILPTL---VRLLHHDDPEVLADTCWAISYLTDGPNERIGMVVKTGVVPQLVK 300
Query: 196 LIKEGKADGQENAARAIG--LLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAV 253
L+ + A RAIG + G D E + +I G +VF +L +Q W +
Sbjct: 301 LLGASELPIVTPALRAIGNIVTGTD-EQTQVVIDAGALAVFPSLLTNPKTNIQKEATWTM 359
Query: 254 SELAANYPKCQELFAQHNIIRLLVGHLA---FETVEE 287
S + A + H ++ LV L+ F+T +E
Sbjct: 360 SNITAGRQDQIQQVVNHGLVPFLVSVLSKADFKTQKE 396
>PTP3_DICDI (P54637) Protein-tyrosine phosphatase 3 (EC 3.1.3.48)
(Protein-tyrosine-phosphate phosphohydrolase 3)
Length = 989
Score = 46.6 bits (109), Expect = 2e-04
Identities = 30/114 (26%), Positives = 44/114 (38%), Gaps = 23/114 (20%)
Query: 311 NNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASKQP 370
NNN+++N N N+D +++TM I +
Sbjct: 103 NNNTTTNNNNNNNNNNDDKFDTNALK--------------------LSNTMIIKNNNNNN 142
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINM 424
N N N N N N+N N N N N N N+ N+NN+ N S+S I +
Sbjct: 143 NNNNNNNNNNN---NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNSNSNIEI 193
Score = 40.8 bits (94), Expect = 0.012
Identities = 31/139 (22%), Positives = 57/139 (40%), Gaps = 7/139 (5%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
+N NNSSSN+N + + + VS + L N +++ +T + +
Sbjct: 56 NNINNSSSNINNNNNNTPDSMSMSTSLSSSPSVSFNHLDLNSIN-NKINNNTTTNNNNNN 114
Query: 369 QPNEGNEAN------QNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGI 422
N ++ + N I+ N+N N N N N N N+ N+NN+ N +++
Sbjct: 115 NNNNDDKFDTNALKLSNTMIIKNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 174
Query: 423 NMKGRESEDAETKASMKEM 441
N + + S E+
Sbjct: 175 NNNNNNNNNNNNSNSNIEI 193
>HYR1_CANAL (P46591) Hyphally regulated protein precursor
Length = 937
Score = 46.6 bits (109), Expect = 2e-04
Identities = 34/126 (26%), Positives = 52/126 (40%), Gaps = 22/126 (17%)
Query: 308 ASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAAS 367
+ +NN S S N G+ + +G G G+ G S G +P
Sbjct: 637 SGSNNGSGSGSNNGSGSGSTEGSEGGSGSNEG--SQSGSGSQPG---------------- 678
Query: 368 KQPNEGNEANQNQNILANSNTPNGNGLGNGNG--NGNDGGKQGNHNNHQRNYSHSGINMK 425
PNEG+E N +N + G+G G+G+G NG+ G Q + ++ S SG N
Sbjct: 679 --PNEGSEGGSGSNEGSNHGSNEGSGSGSGSGSNNGSGSGSQSGSGSGSQSGSESGSNSG 736
Query: 426 GRESED 431
E +
Sbjct: 737 SNEGSN 742
Score = 39.7 bits (91), Expect = 0.027
Identities = 33/135 (24%), Positives = 53/135 (38%), Gaps = 20/135 (14%)
Query: 293 IVSMKANSIHAAVVMASNNNNSSSNLNPKKGTENE------DGVVVGGGNKHGRVSHHPL 346
+V+ N H V + S+ N G E+ +G G GN G S+
Sbjct: 554 VVTYTTNVPHTTVDATTTTTTSTGGDNSTGGNESGSNHGPGNGSTEGSGNGSGAGSNEGS 613
Query: 347 GERPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGK 406
P N S +EG +N + SN +G+G NG+G+G+ G
Sbjct: 614 QSGPNN-------------GSGSGSEGG-SNNGSGSDSGSNNGSGSGSNNGSGSGSTEGS 659
Query: 407 QGNHNNHQRNYSHSG 421
+G +++ + S SG
Sbjct: 660 EGGSGSNEGSQSGSG 674
Score = 37.7 bits (86), Expect = 0.10
Identities = 26/114 (22%), Positives = 45/114 (38%), Gaps = 4/114 (3%)
Query: 310 NNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASKQ 369
+ S SN G+ G G G+ +G S G + + + +
Sbjct: 683 SEGGSGSNEGSNHGSNEGSGSGSGSGSNNGSGSGSQSGSGSGSQSGSESGSNSGSNEGSN 742
Query: 370 PNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGIN 423
P GN +N+ + + NG+ G+G G+G + G HN+ + S+ G N
Sbjct: 743 PGAGNGSNEG----SGQGSGNGSEAGSGQGSGPNNGSGSGHNDGSGSGSNQGSN 792
Score = 33.5 bits (75), Expect = 1.9
Identities = 30/128 (23%), Positives = 47/128 (36%), Gaps = 28/128 (21%)
Query: 308 ASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAAS 367
+ +N+ S+ NP G + +G G GN S G
Sbjct: 731 SGSNSGSNEGSNPGAGNGSNEGSGQGSGNGSEAGSGQGSG-------------------- 770
Query: 368 KQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGR 427
PN G+ + N + SN + G G+G+G+ G K G+H+ S+ G
Sbjct: 771 --PNNGSGSGHNDGSGSGSNQGSNPGAGSGSGS-ESGSKAGSHSG-----SNEGAKTDSI 822
Query: 428 ESEDAETK 435
E E+K
Sbjct: 823 EGFHTESK 830
Score = 33.1 bits (74), Expect = 2.5
Identities = 24/80 (30%), Positives = 32/80 (40%), Gaps = 5/80 (6%)
Query: 358 TSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNY 417
T+T GNE+ N N +T G GNG+G G++ G Q NN +
Sbjct: 569 TTTTTTSTGGDNSTGGNESGSNHGP-GNGST---EGSGNGSGAGSNEGSQSGPNNGSGSG 624
Query: 418 SHSGINMKGRESEDAETKAS 437
S G N G S+ S
Sbjct: 625 SEGGSN-NGSGSDSGSNNGS 643
Score = 31.2 bits (69), Expect = 9.5
Identities = 16/59 (27%), Positives = 25/59 (42%)
Query: 373 GNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESED 431
GN + N + S NG+G G+ G+ N G NN + S++G E +
Sbjct: 602 GNGSGAGSNEGSQSGPNNGSGSGSEGGSNNGSGSDSGSNNGSGSGSNNGSGSGSTEGSE 660
>OMB_DROME (Q24432) Optomotor-blind protein
(Lethal(1)optomotor-blind) (L(1)omb) (Bifid protein)
Length = 972
Score = 44.3 bits (103), Expect = 0.001
Identities = 32/106 (30%), Positives = 44/106 (41%), Gaps = 21/106 (19%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVV----GGGNKHGRVSHHPLGERPRNLHRVITSTMAIH 364
S NNNS+SN N T N + +V GGG + S+H T+T +
Sbjct: 60 SGNNNSNSNNNTNSNTNNTNNLVAVSPTGGGAQLSPQSNHSSSN---------TTTTS-- 108
Query: 365 AASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNH 410
N N ++ N N + N N + N N N N KQG+H
Sbjct: 109 ------NTNNSSSNNNNNNSTHNNNNNHTNNNNNNNNNTSQKQGHH 148
Score = 38.5 bits (88), Expect = 0.060
Identities = 24/116 (20%), Positives = 47/116 (39%), Gaps = 16/116 (13%)
Query: 303 AAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMA 362
++++ A +NNN+S N N N S++ N + ++ +
Sbjct: 43 SSLLTAGSNNNNSGNTNSGNNNSN---------------SNNNTNSNTNNTNNLVAVSPT 87
Query: 363 IHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYS 418
A P + N ++ N +N+N + N N + + N+ N+NN+ N S
Sbjct: 88 GGGAQLSP-QSNHSSSNTTTTSNTNNSSSNNNNNNSTHNNNNNHTNNNNNNNNNTS 142
>ECR_LUCCU (O18531) Ecdysone receptor (Ecdysteroid receptor)
(20-hydroxy-ecdysone receptor) (20E receptor)
Length = 757
Score = 43.9 bits (102), Expect = 0.001
Identities = 46/182 (25%), Positives = 70/182 (38%), Gaps = 29/182 (15%)
Query: 278 GHLAFETVEEHSKYAIVSMKANSIHAAVVMASNNNNSSSNLN-PKKGTENEDGVVVGGGN 336
G A + +EE S + S +V++S+ N S S+L+ P G +++ +
Sbjct: 11 GFAALKMLEESSS------EVTSSSNGLVLSSDINMSPSSLDSPVYG--DQEMWLCNDSA 62
Query: 337 KHGRVSHHPLGERPRNLHRVITSTMAIHAASKQPNEGNEA--NQNQN----------ILA 384
+ H + + + + I S PN N + NQNQN +
Sbjct: 63 SYNNSHQHSVITSLQGCTSSLPAQTTIIPLSALPNSNNASLNNQNQNYQNGNSMNTNLSV 122
Query: 385 NSNTPNGNGLGNG--------NGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKA 436
N+N G G G G NG G GG Q N++NH N+ H N S T
Sbjct: 123 NTNNSVGGGGGGGGVPGMTSLNGLGGGGGSQVNNHNHSHNHLHHNSNSNHSNSSSHHTNG 182
Query: 437 SM 438
M
Sbjct: 183 HM 184
>P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K)
Length = 1858
Score = 43.5 bits (101), Expect = 0.002
Identities = 22/66 (33%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query: 366 ASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGN--HNNHQRNYSHSGIN 423
++K+ + + N N N N+N N N N N N N+G GN +NN N S I+
Sbjct: 1005 SNKENKDSSSNNNNNNNNNNNNNNNNNNNNNNNNNNNNGNNNGNNSNNNSNSNISRGSID 1064
Query: 424 MKGRES 429
+G S
Sbjct: 1065 SEGNGS 1070
Score = 42.0 bits (97), Expect = 0.005
Identities = 23/82 (28%), Positives = 39/82 (47%), Gaps = 1/82 (1%)
Query: 357 ITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRN 416
I + + I +K+ + N+ N++ + N+N N N N N N N+ N+ N+ N
Sbjct: 989 IQTVINIKETNKENKDSNKENKDSSSNNNNNNNNNNNNNNNNNNNNNNNNNNNNGNNNGN 1048
Query: 417 YSHSGINMK-GRESEDAETKAS 437
S++ N R S D+E S
Sbjct: 1049 NSNNNSNSNISRGSIDSEGNGS 1070
Score = 39.7 bits (91), Expect = 0.027
Identities = 18/59 (30%), Positives = 27/59 (45%)
Query: 358 TSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRN 416
+S+ + A + P + N N N N+N N N N N N N+ N+NN+ N
Sbjct: 168 SSSSSTTATTPSPTTTSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 226
Score = 39.3 bits (90), Expect = 0.035
Identities = 19/71 (26%), Positives = 33/71 (45%), Gaps = 1/71 (1%)
Query: 346 LGERPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGG 405
+G +P + + +S+ + + P+ +N N N N+N N N N N N N+
Sbjct: 155 IGSKPI-CNNLTSSSSSSSTTATTPSPTTTSNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 213
Query: 406 KQGNHNNHQRN 416
N+NN+ N
Sbjct: 214 NNNNNNNNNNN 224
Score = 38.5 bits (88), Expect = 0.060
Identities = 19/77 (24%), Positives = 31/77 (39%)
Query: 336 NKHGRVSHHPLGERPRNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLG 395
++ G + P+ + ++T + + N N N N N N+N N N
Sbjct: 150 SRSGSIGSKPICNNLTSSSSSSSTTATTPSPTTTSNNNNNNNNNNNNNNNNNNNNNNNNN 209
Query: 396 NGNGNGNDGGKQGNHNN 412
N N N N+ N+NN
Sbjct: 210 NNNNNNNNNNNNNNNNN 226
Score = 37.0 bits (84), Expect = 0.17
Identities = 19/71 (26%), Positives = 34/71 (47%), Gaps = 3/71 (4%)
Query: 368 KQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGR 427
K+ N+ N+ + +N ++SN N N N N N N+ N+NN+ N ++ N
Sbjct: 996 KETNKENKDSNKENKDSSSNNNNNN---NNNNNNNNNNNNNNNNNNNNNNGNNNGNNSNN 1052
Query: 428 ESEDAETKASM 438
S ++ S+
Sbjct: 1053 NSNSNISRGSI 1063
Score = 35.0 bits (79), Expect = 0.66
Identities = 18/90 (20%), Positives = 37/90 (41%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
SNNNN+++N N N + N + +++ T+T + ++S
Sbjct: 184 SNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTSTTTTTTSILISSSP 243
Query: 369 QPNEGNEANQNQNILANSNTPNGNGLGNGN 398
P+ + ++ N N+N N + G +
Sbjct: 244 PPSSSSSSSSNDEQFNNNNNNNNSNSGGSS 273
Score = 34.7 bits (78), Expect = 0.86
Identities = 15/51 (29%), Positives = 23/51 (44%)
Query: 384 ANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAET 434
+N+N N N N N N N+ N+NN+ N +++ N S T
Sbjct: 184 SNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTSTTTTT 234
Score = 34.3 bits (77), Expect = 1.1
Identities = 21/97 (21%), Positives = 40/97 (40%), Gaps = 8/97 (8%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
++NNN+++N N N + N + +++ N T+T +I +S
Sbjct: 183 TSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTSTTTTTTSILISSS 242
Query: 369 QPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGG 405
P + ++ + + N+N N N N N GG
Sbjct: 243 PPPSSSSSSSSNDEQFNNN--------NNNNNSNSGG 271
Score = 33.5 bits (75), Expect = 1.9
Identities = 14/54 (25%), Positives = 24/54 (43%)
Query: 385 NSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASM 438
++N N N N N N N+ N+NN+ N +++ N + T S+
Sbjct: 184 SNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNTTSTTTTTTSI 237
Score = 32.7 bits (73), Expect = 3.3
Identities = 17/51 (33%), Positives = 23/51 (44%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSG 421
N N N N N N+N NGN GN + N ++ + + N S SG
Sbjct: 1023 NNNNNNNNNNNNNNNNNNNNGNNNGNNSNNNSNSNISRGSIDSEGNGSGSG 1073
Score = 31.2 bits (69), Expect = 9.5
Identities = 14/37 (37%), Positives = 19/37 (50%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQ 407
N GN N N +NSN G+ GNG+G+ G +
Sbjct: 1041 NNGNNNGNNSNNNSNSNISRGSIDSEGNGSGSGNGSE 1077
Score = 31.2 bits (69), Expect = 9.5
Identities = 12/38 (31%), Positives = 19/38 (49%)
Query: 386 SNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGIN 423
+ T N N N N N N+ N+NN+ N +++ N
Sbjct: 181 TTTSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 218
>MAM_DROME (P21519) Neurogenic protein mastermind
Length = 1596
Score = 43.5 bits (101), Expect = 0.002
Identities = 35/145 (24%), Positives = 52/145 (35%), Gaps = 34/145 (23%)
Query: 307 MASNNNNSSSNLNPKKGTENEDGVVV----------------GGGNKHG----------- 339
+ +NNNNS+SN N N G GG N +G
Sbjct: 257 LPNNNNNSNSNNNNGNANANNGGNGSNTGNNTNNNGNSTNNNGGSNNNGSENLTKFSVEI 316
Query: 340 ----RVSHHPLGERPRNLHRVITSTMAIHAASK-QPNEGNEANQNQNILANSNTPNGNGL 394
+ P +P+ + +T + + K +P G NS N NG
Sbjct: 317 VQQLEFTTSPANSQPQQISTNVTVKALTNTSVKSEPGVGGGGGGGGG--GNSGNNNNNGG 374
Query: 395 GNGNGNGNDGGKQGNHNNHQRNYSH 419
G G GNGN+ G+H+ Q+ + H
Sbjct: 375 GGGGGNGNNNNNGGDHHQQQQQHQH 399
Score = 40.4 bits (93), Expect = 0.016
Identities = 22/62 (35%), Positives = 30/62 (47%), Gaps = 9/62 (14%)
Query: 368 KQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGR 427
K PN N +N N N NGN N GNG++ G N+N + N ++ G N G
Sbjct: 256 KLPNNNNNSNSNNN--------NGNANANNGGNGSNTGNNTNNNGNSTN-NNGGSNNNGS 306
Query: 428 ES 429
E+
Sbjct: 307 EN 308
>IMA2_XENLA (P52171) Importin alpha-2 subunit (Karyopherin alpha-2
subunit)
Length = 522
Score = 43.5 bits (101), Expect = 0.002
Identities = 25/104 (24%), Positives = 53/104 (50%), Gaps = 1/104 (0%)
Query: 152 IAMLFTGSQEVRSDAAASLVSLARGSDRYGKLIIEEGGVGPLLKLIKEGKADGQENAARA 211
I ++++ + + + ++ ++ G+D+ + I+ G + L +L++ K Q+ AA A
Sbjct: 291 IQLMYSPELSILTPSLRTVGNIVTGTDKQTQAAIDAGVLSVLPQLLRHQKPSIQKEAAWA 350
Query: 212 IG-LLGRDAESVEHMIHVGVCSVFAKILKEGPMKVQGVVAWAVS 254
+ + A ++ MI G+ S +LK+G K Q WAV+
Sbjct: 351 LSNIAAGPAPQIQQMITCGLLSPLVDLLKKGDFKAQKEAVWAVT 394
>IF2_LISIN (Q92C29) Translation initiation factor IF-2
Length = 782
Score = 42.7 bits (99), Expect = 0.003
Identities = 35/113 (30%), Positives = 44/113 (37%), Gaps = 16/113 (14%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAASK 368
+N+N S+ N G N+ HG P +P N TS + +K
Sbjct: 81 TNSNEKSNKPNKPAGQANKPATA---NKSHGA---KPATNKPAN-----TSNQTQSSGNK 129
Query: 369 QPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSG 421
QP G + N N N SN P G G N G N + GN NN RN G
Sbjct: 130 QPAGGQKRNNNNN----SNRPGG-GNPNRPGGNNRPNRGGNFNNKGRNTKKKG 177
Score = 34.3 bits (77), Expect = 1.1
Identities = 39/170 (22%), Positives = 63/170 (36%), Gaps = 23/170 (13%)
Query: 282 FETVEEH---SKYAIVSMKANSIHAAVVMASNNNNSSSNLN------------PKKGTEN 326
+E +EH SK I ++K I A M++ N N+ L+ PKK T +
Sbjct: 7 YEYAKEHQVSSKKVIEALKDLGIEVANHMSTINENALRQLDNAIDGTNKKAEAPKKETTS 66
Query: 327 EDGVVVGGGNKHGRVSHHPLGERPRN----LHRVITSTMAIHAASKQPNEGNEANQNQNI 382
+ G NK + + +P ++ T+ + A N +NQ Q+
Sbjct: 67 NENGNSKGPNKPNMTNSNEKSNKPNKPAGQANKPATANKSHGAKPATNKPANTSNQTQS- 125
Query: 383 LANSNTPNGNGLGNGNGNG--NDGGKQGNHNNHQRNYSHSGINMKGRESE 430
+ + P G N N N GG + R N KGR ++
Sbjct: 126 -SGNKQPAGGQKRNNNNNSNRPGGGNPNRPGGNNRPNRGGNFNNKGRNTK 174
>PI4K_DICDI (P54677) Phosphatidylinositol 4-kinase (EC 2.7.1.67)
(PI4-kinase) (PtdIns-4-kinase) (PI4K-alpha)
Length = 1093
Score = 42.4 bits (98), Expect = 0.004
Identities = 26/122 (21%), Positives = 54/122 (43%), Gaps = 4/122 (3%)
Query: 306 VMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVIT---STMA 362
+ ++NNNN++ N N +N + ++ N ++ N + I+ + +
Sbjct: 183 INSNNNNNNNINNNNSNNDDNNNNEILPNENSDNSINDENNQYGNSNNNNNISGENNNIK 242
Query: 363 IHAASKQPNEGNEANQNQNILANSNT-PNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSG 421
I S+ ++ N N + + T P + + N N N N+ N+NN+ N +++
Sbjct: 243 IDINSQNKSDSNIETLNSTLCEETKTSPIKDDMENNNNNNNNNNNNNNNNNNNNNINNNN 302
Query: 422 IN 423
IN
Sbjct: 303 IN 304
Score = 41.6 bits (96), Expect = 0.007
Identities = 30/113 (26%), Positives = 51/113 (44%), Gaps = 10/113 (8%)
Query: 310 NNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAIHAAS-- 367
N++NS ++ N + G N + + G N + ++ + + N+ + ST+ +
Sbjct: 213 NSDNSINDENNQYGNSNNNNNI-SGENNNIKIDINSQNKSDSNIE-TLNSTLCEETKTSP 270
Query: 368 -KQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSH 419
K E N N N N N+N N N + N N N N+ +NN+ NY H
Sbjct: 271 IKDDMENNNNNNNNNNNNNNNNNNNNNINNNNINNNN-----INNNNNINYGH 318
Score = 34.7 bits (78), Expect = 0.86
Identities = 28/121 (23%), Positives = 52/121 (42%), Gaps = 7/121 (5%)
Query: 309 SNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHP--LGERPR---NLHRVITSTMAI 363
+N+NN +N N EN D + N++G +++ GE +++ S I
Sbjct: 196 NNSNNDDNNNNEILPNENSDNSINDENNQYGNSNNNNNISGENNNIKIDINSQNKSDSNI 255
Query: 364 HAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGIN 423
+ E + + ++ + N+N N N N N N N+ N+NN N ++ N
Sbjct: 256 ETLNSTLCEETKTSPIKDDMENNNNNNNNN--NNNNNNNNNNNNINNNNINNNNINNNNN 313
Query: 424 M 424
+
Sbjct: 314 I 314
Score = 31.6 bits (70), Expect = 7.3
Identities = 20/66 (30%), Positives = 27/66 (40%), Gaps = 7/66 (10%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNN--HQRNYSHSGINMKGRE 428
N N+ N N IL N N+ N N N G N+NN + N IN + +
Sbjct: 197 NSNNDDNNNNEILPNENSDNSI-----NDENNQYGNSNNNNNISGENNNIKIDINSQNKS 251
Query: 429 SEDAET 434
+ ET
Sbjct: 252 DSNIET 257
>P3K3_DICDI (P54675) Phosphatidylinositol 3-kinase 3 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K) (Fragment)
Length = 1585
Score = 42.4 bits (98), Expect = 0.004
Identities = 40/169 (23%), Positives = 64/169 (37%), Gaps = 39/169 (23%)
Query: 309 SNNNNSSSNLNPK---KGTENEDGVVVGGGNKH------------------------GRV 341
+NNNN+S+N N T N G + N + G
Sbjct: 241 NNNNNNSNNTNSNNTTNATTNSVGFSITMTNSNSLSVSKRMNKFKSWTSSKPTSSSIGFA 300
Query: 342 SHHPLGERPRNL---HRVITS------TMAIHAASKQPNEGNEANQNQNILANSNTPNGN 392
S +P N+ R TS + +S P + + N+N N N+N N N
Sbjct: 301 SSPQNNGKPLNISGSSRFFTSRQDSKIDLLKSPSSSPPTQSDIFNENNN---NNNNNNNN 357
Query: 393 GLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESEDAETKASMKEM 441
N N N N+ N+NN++ +++ N + ET+ S+KE+
Sbjct: 358 NNNNNNNNNNNNNNNNNNNNNEELINNNNNNNNDENYKIEETEESLKEL 406
Score = 36.6 bits (83), Expect = 0.23
Identities = 32/134 (23%), Positives = 47/134 (34%), Gaps = 40/134 (29%)
Query: 309 SNNNNSSSNLNPKKG-------TENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTM 361
++NNN++ NL+P TEN+ + K PL + P +
Sbjct: 102 NSNNNNNINLSPDSSITKDINITENKITEIKTTETKETSTGTSPLEKSP---------SK 152
Query: 362 AIHAASKQPNEGNEANQN------------------------QNILANSNTPNGNGLGNG 397
+ K+P E NE +NI N+N N NG GN
Sbjct: 153 GFIISPKKPEEENEIEGETINNIAITNYTQGPSMLTLMKKKLENIKKNNNNNNNNGNGNN 212
Query: 398 NGNGNDGGKQGNHN 411
N N N+ N+N
Sbjct: 213 NSNNNNSNSNNNNN 226
Score = 34.3 bits (77), Expect = 1.1
Identities = 17/61 (27%), Positives = 24/61 (38%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESE 430
N N +N I N N N N N N N+ N+NN+ N + + +E
Sbjct: 39 NSLNNSNIYLTIPTTQNLINNNNNNNNNNNNNNNNNNNNNNNNNNNVIIPSASTENKEEN 98
Query: 431 D 431
D
Sbjct: 99 D 99
Score = 32.7 bits (73), Expect = 3.3
Identities = 18/66 (27%), Positives = 28/66 (42%), Gaps = 11/66 (16%)
Query: 368 KQPNEGNEANQNQNILANSNTPNGNGLGNG-----------NGNGNDGGKQGNHNNHQRN 416
K+ N N N N N +N+N N N NG NGN N+ ++N+ N
Sbjct: 198 KKNNNNNNNNGNGNNNSNNNNSNSNNNNNGISPSSSPPSHLNGNNNNNNSNNTNSNNTTN 257
Query: 417 YSHSGI 422
+ + +
Sbjct: 258 ATTNSV 263
Score = 32.7 bits (73), Expect = 3.3
Identities = 17/62 (27%), Positives = 29/62 (46%), Gaps = 5/62 (8%)
Query: 377 NQNQNILANSNT-----PNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESED 431
N +N L NSN N + N N N N+ N+NN+ N +++ + + +E+
Sbjct: 35 NIKENSLNNSNIYLTIPTTQNLINNNNNNNNNNNNNNNNNNNNNNNNNNNVIIPSASTEN 94
Query: 432 AE 433
E
Sbjct: 95 KE 96
Score = 32.0 bits (71), Expect = 5.6
Identities = 19/70 (27%), Positives = 29/70 (41%), Gaps = 8/70 (11%)
Query: 370 PNEGNEANQNQNILANSNTPNGNGLGNGNGNGND--------GGKQGNHNNHQRNYSHSG 421
P N N N N N+N N N N N N N+ K+ N NN+ N ++
Sbjct: 51 PTTQNLINNNNNNNNNNNNNNNNNNNNNNNNNNNVIIPSASTENKEENDNNNSNNNNNIN 110
Query: 422 INMKGRESED 431
++ ++D
Sbjct: 111 LSPDSSITKD 120
>GPR1_YEAST (Q12361) G protein-coupled receptor GPR1
Length = 961
Score = 42.4 bits (98), Expect = 0.004
Identities = 20/77 (25%), Positives = 34/77 (43%)
Query: 366 ASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMK 425
++ ++ N N N N NSN N N N N N N+ N+NN+ N ++ N+
Sbjct: 494 SNNDDDDNNNNNDNDNDNNNSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNSNNIKNNVD 553
Query: 426 GRESEDAETKASMKEMA 442
+ A+ ++ A
Sbjct: 554 NNNTNPADNIPTLSNEA 570
Score = 37.4 bits (85), Expect = 0.13
Identities = 19/70 (27%), Positives = 31/70 (44%)
Query: 351 RNLHRVITSTMAIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNH 410
+N + ITS + N N+ + N N N N N + N N N N+ N+
Sbjct: 473 KNDNSDITSNIKEKGGIINNNSNNDDDDNNNNNDNDNDNNNSNNNNNNNNNNNNNNNNNN 532
Query: 411 NNHQRNYSHS 420
NN+ N +++
Sbjct: 533 NNNNNNNNNN 542
>VLPA_MYCHR (P29228) Variant surface antigen A precursor (VlpA
prolipoprotein)
Length = 176
Score = 42.0 bits (97), Expect = 0.005
Identities = 24/135 (17%), Positives = 55/135 (39%), Gaps = 10/135 (7%)
Query: 304 AVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTMAI 363
A+ +NNSS + P GT N G G+ +G + G ++ +
Sbjct: 27 AISCGQTDNNSSQSQQPGSGTTNTSGGTNSSGSTNGTAGTNSSG----------STNGSG 76
Query: 364 HAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGIN 423
+ ++ + N GN+ N +++ + G G+G+ ++ G + + G N
Sbjct: 77 NGSNSETNTGNKTTSESNSGSSTGSQAGTTTNTGSGSNSESGMNSEKTENTQQSEAPGTN 136
Query: 424 MKGRESEDAETKASM 438
+ + ++ +++ M
Sbjct: 137 TGNKTTSESNSESGM 151
>YIN0_YEAST (P40467) Putative 108.8 kDa transcriptional regulatory
protein in FKH1-STH1 intergenic region
Length = 964
Score = 41.6 bits (96), Expect = 0.007
Identities = 30/124 (24%), Positives = 49/124 (39%), Gaps = 31/124 (25%)
Query: 302 HAAVVMASNNNNSSSNLNPKKGTENEDGVVVGGGNKHGRVSHHPLGERPRNLHRVITSTM 361
++A +S+ +N+S+N NP N + V G N + +++
Sbjct: 800 NSAFDFSSSKSNASNNSNPDTINNNYNNVS-GKNNNNNNITN------------------ 840
Query: 362 AIHAASKQPNEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSG 421
N N N N N N+N N N N N N N G N+NN+ N +++
Sbjct: 841 ---------NSNNNHNNNNNDNNNNNNNNNN---NNNNNNNSGNSSNNNNNNNNNKNNND 888
Query: 422 INMK 425
+K
Sbjct: 889 FGIK 892
Score = 40.8 bits (94), Expect = 0.012
Identities = 22/80 (27%), Positives = 35/80 (43%), Gaps = 5/80 (6%)
Query: 357 ITSTMAIHAASKQPNEGNEAN-----QNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHN 411
I + A +S + N N +N N N ++ N N N N N N N+ N+N
Sbjct: 797 IPTNSAFDFSSSKSNASNNSNPDTINNNYNNVSGKNNNNNNITNNSNNNHNNNNNDNNNN 856
Query: 412 NHQRNYSHSGINMKGRESED 431
N+ N +++ N G S +
Sbjct: 857 NNNNNNNNNNNNNSGNSSNN 876
Score = 40.0 bits (92), Expect = 0.020
Identities = 18/61 (29%), Positives = 29/61 (47%)
Query: 371 NEGNEANQNQNILANSNTPNGNGLGNGNGNGNDGGKQGNHNNHQRNYSHSGINMKGRESE 430
N + N N NI NSN + N + N N N+ N+NN+ N S++ N ++
Sbjct: 827 NVSGKNNNNNNITNNSNNNHNNNNNDNNNNNNNNNNNNNNNNNSGNSSNNNNNNNNNKNN 886
Query: 431 D 431
+
Sbjct: 887 N 887
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.131 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 76,268,903
Number of Sequences: 164201
Number of extensions: 3205165
Number of successful extensions: 16019
Number of sequences better than 10.0: 203
Number of HSP's better than 10.0 without gapping: 86
Number of HSP's successfully gapped in prelim test: 125
Number of HSP's that attempted gapping in prelim test: 12195
Number of HSP's gapped (non-prelim): 1293
length of query: 687
length of database: 59,974,054
effective HSP length: 117
effective length of query: 570
effective length of database: 40,762,537
effective search space: 23234646090
effective search space used: 23234646090
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147013.12


