

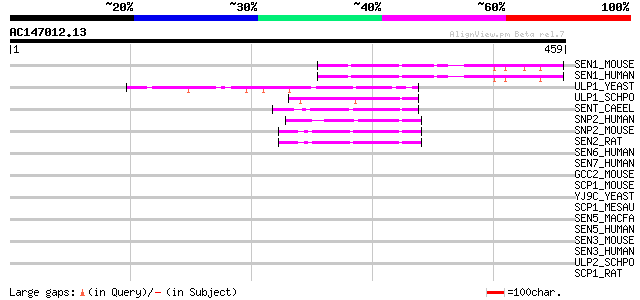
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147012.13 + phase: 0 /pseudo
(459 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SEN1_MOUSE (P59110) Sentrin-specific protease 1 (EC 3.4.22.-) (S... 64 1e-09
SEN1_HUMAN (Q9P0U3) Sentrin-specific protease 1 (EC 3.4.22.-) (S... 61 7e-09
ULP1_YEAST (Q02724) Ubiquitin-like-specific protease 1 (EC 3.4.2... 57 1e-07
ULP1_SCHPO (O42957) Ubiquitin-like-specific protease 1 (EC 3.4.2... 54 8e-07
SENT_CAEEL (Q09353) Probable sentrin-specific protease (EC 3.4.2... 54 1e-06
SNP2_HUMAN (Q9HC62) Sentrin-specific protease 2 (EC 3.4.22.-) (S... 51 7e-06
SNP2_MOUSE (Q91ZX6) Sentrin-specific protease 2 (EC 3.4.22.-) (S... 49 3e-05
SEN2_RAT (Q9EQE1) Sentrin-specific protease 2 (EC 3.4.22.-) (Sen... 49 3e-05
SEN6_HUMAN (Q9GZR1) Sentrin-specific protease 6 (EC 3.4.22.-) (S... 43 0.001
SEN7_HUMAN (Q9BQF6) Sentrin-specific protease 7 (EC 3.4.22.-) (S... 40 0.010
GCC2_MOUSE (Q8CHG3) GRIP and coiled-coil domain-containing prote... 40 0.013
SCP1_MOUSE (Q62209) Synaptonemal complex protein 1 (SCP-1 protein) 39 0.036
YJ9C_YEAST (P47166) Hypothetical 81.2 kDa protein in NMD5-HOM6 i... 38 0.048
SCP1_MESAU (Q60563) Synaptonemal complex protein 1 (SCP-1 protei... 38 0.048
SEN5_MACFA (Q8WP32) Sentrin-specific protease 5 (EC 3.4.22.-) (S... 37 0.081
SEN5_HUMAN (Q96HI0) Sentrin-specific protease 5 (EC 3.4.22.-) (S... 37 0.081
SEN3_MOUSE (Q9EP97) Sentrin-specific protease 3 (EC 3.4.22.-) (S... 37 0.081
SEN3_HUMAN (Q9H4L4) Sentrin-specific protease 3 (EC 3.4.22.-) (S... 37 0.081
ULP2_SCHPO (O13769) Ubiquitin-like-specific protease 2 (EC 3.4.2... 37 0.11
SCP1_RAT (Q03410) Synaptonemal complex protein 1 (SCP-1 protein) 37 0.14
>SEN1_MOUSE (P59110) Sentrin-specific protease 1 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP1)
Length = 640
Score = 63.5 bits (153), Expect = 1e-09
Identities = 63/243 (25%), Positives = 107/243 (43%), Gaps = 57/243 (23%)
Query: 255 EPFVPLTKEEEVEVTRAF-SANQNKVLVNHEKSNIEISGKMFRCLRPGEWLNDEVINLYL 313
+ F +T+E E E+ F + NQ++VL E + I+ K + L WLNDE+IN Y+
Sbjct: 414 DEFPEITEEMEKEIKNVFRNGNQDEVL--SEAFRLTITRKDIQTLNHLNWLNDEIINFYM 471
Query: 314 ELLKERERREPQKFLKCHFFNTFFYKKQECL*LQISEEMDYSKEIGIWPT*M**NICTYS 373
+L ER + + F H FNTFF+ K + Q + ++K++ ++
Sbjct: 472 NMLMERSKE--KGFPSVHAFNTFFFTKLKTAGYQAVKR--WTKKVDVFSV---------- 517
Query: 374 PGDPLVLSCYQ*EGKEISVS*FTERN--*HRSAGSAD-------------EVKDKTGEDV 418
D L++ + ++V F ++ + S G + E DK ++
Sbjct: 518 --DILLVPIHLGVHWCLAVVDFRRKSITYYDSMGGINNEACRILLQYLKQESVDKKRKEF 575
Query: 419 DISSWE--TEFVEDLPEQMNG---------------------FVIEHMPYFRLRTAKEIL 455
D + W+ ++ +++P+QMNG F +HMPYFR R EIL
Sbjct: 576 DTNGWQLFSKKSQEIPQQMNGSDCGMFACKYADCITKDRPINFTQQHMPYFRKRMVWEIL 635
Query: 456 RLK 458
K
Sbjct: 636 HRK 638
>SEN1_HUMAN (Q9P0U3) Sentrin-specific protease 1 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP1)
Length = 643
Score = 60.8 bits (146), Expect = 7e-09
Identities = 63/242 (26%), Positives = 103/242 (42%), Gaps = 56/242 (23%)
Query: 255 EPFVPLTKEEEVEVTRAF-SANQNKVLVNHEKSNIEISGKMFRCLRPGEWLNDEVINLYL 313
+ F +T+E E E+ F + NQ++VL E + I+ K + L WLNDE+IN Y+
Sbjct: 418 DEFPEITEEMEKEIKNVFRNGNQDEVL--SEAFRLTITRKDIQTLNHLNWLNDEIINFYM 475
Query: 314 ELLKERERREPQKFLKCHFFNTFFYKKQECL*LQISEEMDYSKEIGIWPT*M**NICTYS 373
+L ER + + H FNTFF+ K + Q + ++K++ ++
Sbjct: 476 NMLMERSKE--KGLPSVHAFNTFFFTKLKTAGYQAVKR--WTKKVDVFSV---------- 521
Query: 374 PGDPLVLSCYQ*EGKEISVS*FTERN--*HRSAGSAD-------------EVKDKTGEDV 418
D L++ + ++V F ++N + S G + E DK ++
Sbjct: 522 --DILLVPIHLGVHWCLAVVDFRKKNITYYDSMGGINNEACRILLQYLKQESIDKKRKEF 579
Query: 419 DISSWET-EFVEDLPEQMNG---------------------FVIEHMPYFRLRTAKEILR 456
D + W+ +P+QMNG F +HMPYFR R EIL
Sbjct: 580 DTNGWQLFSKKSQIPQQMNGSDCGMFACKYADCITKDRPINFTQQHMPYFRKRMVWEILH 639
Query: 457 LK 458
K
Sbjct: 640 RK 641
>ULP1_YEAST (Q02724) Ubiquitin-like-specific protease 1 (EC
3.4.22.-)
Length = 621
Score = 56.6 bits (135), Expect = 1e-07
Identities = 62/278 (22%), Positives = 126/278 (45%), Gaps = 59/278 (21%)
Query: 97 ASDTTDELMEKYEHAKHAALATISFREKGKVKDNIKEVVVLKDSNVEDVE---------- 146
+ + T + Y+ ++ T R+K K NI ++ + E+V
Sbjct: 222 SENNTSDQKNSYDRRQYG---TAFIRKKKVAKQNINNTKLVSRAQSEEVTYLRQIFNGEY 278
Query: 147 EFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKMLDNL-------SLSTK 199
+ ++K+ + ++ L+ +D + + G+++ S++D + + + L T+
Sbjct: 279 KVPKILKEERERQLKLMDMDKEKDT---GLKK----SIIDLTEKIKTILIENNKNRLQTR 331
Query: 200 NDLSGVCAY---KKLLEAVDKRGDTLG-RLKFEIQ--------------LNEKRKSAFDL 241
N+ + KK+ K D L +LKF+ LNE++K DL
Sbjct: 332 NENDDDLVFVKEKKISSLERKHKDYLNQKLKFDRSILEFEKDFKRYNEILNERKKIQEDL 391
Query: 242 LRPKKELVEEVPREPFVP-LTKEEEVEVTRAFSANQNKVLVNHEKSNIEISGKMFRCLRP 300
+ K++L ++ VP L ++++ +V +A ++ +N L+N + NIEI+ + F+ L P
Sbjct: 392 KKKKEQLA----KKKLVPELNEKDDDQVQKALASRENTQLMNRD--NIEITVRDFKTLAP 445
Query: 301 GEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFY 338
WLND +I +++ + E+ P FN+FFY
Sbjct: 446 RRWLNDTIIEFFMKYI---EKSTPNTVA----FNSFFY 476
>ULP1_SCHPO (O42957) Ubiquitin-like-specific protease 1 (EC
3.4.22.-)
Length = 568
Score = 53.9 bits (128), Expect = 8e-07
Identities = 42/120 (35%), Positives = 59/120 (49%), Gaps = 13/120 (10%)
Query: 231 LNEKRKSAF------DLLRPKKELVEEVPREP-FVPLTKEEEVEVTRAFS-ANQNKVLVN 282
L EK+ +F D L+ K + P P F+P + + RA NQ+ + +
Sbjct: 308 LKEKQTESFQDWNEVDFLQLKGLEISPPPTRPKFIPELEFPDNARKRALKYLNQSNSVSS 367
Query: 283 HE----KSNIEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFY 338
E K NI I+ K LR +WLNDEVIN Y+ L+ ER + + + H FNTFFY
Sbjct: 368 SEPIITKFNIPITLKDLHTLRNRQWLNDEVINFYMNLISERSKID-SSLPRVHGFNTFFY 426
>SENT_CAEEL (Q09353) Probable sentrin-specific protease (EC
3.4.22.-)
Length = 697
Score = 53.5 bits (127), Expect = 1e-06
Identities = 42/122 (34%), Positives = 63/122 (51%), Gaps = 13/122 (10%)
Query: 218 RGDTLGRLKFEIQLNEKRKSAFDLLRPKKELVEEVPREPFVPLTKEEEVEVTRAFSA-NQ 276
RGD L ++ ++L +RPK VE+ + F+ L + V RA+S N
Sbjct: 441 RGDRLEDVRKRLELQGIA------IRPK---VEKKKVDDFMALPDAADALVERAWSGGNP 491
Query: 277 NKVLVNHEKSNIEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTF 336
N+ V + +I+I K L WLNDE+IN YL+L+ +R + K+ K + FNTF
Sbjct: 492 NEQFV--DAFSIQICKKDLATLSGLHWLNDEIINFYLQLICDRSNGD-SKYPKIYAFNTF 548
Query: 337 FY 338
FY
Sbjct: 549 FY 550
>SNP2_HUMAN (Q9HC62) Sentrin-specific protease 2 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP2) (SMT3-specific
isopeptidase 2) (Smt3ip2) (Axam2)
Length = 589
Score = 50.8 bits (120), Expect = 7e-06
Identities = 38/113 (33%), Positives = 59/113 (51%), Gaps = 14/113 (12%)
Query: 229 IQLNEKRKSAFDLLRPKKELVEEVPREPFVPLTKEEEVEVTRAFSAN-QNKVLVNHEKSN 287
I+ EK S + R +L+E LT++ E E++ A Q+++L + K
Sbjct: 346 IETKEKNCSGKERDRRTDDLLE---------LTEDMEKEISNALGHGPQDEILSSAFK-- 394
Query: 288 IEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
+ I+ + L+ WLNDEVIN Y+ LL ER ++ Q + H F+TFFY K
Sbjct: 395 LRITRGDIQTLKNYHWLNDEVINFYMNLLVERNKK--QGYPALHVFSTFFYPK 445
>SNP2_MOUSE (Q91ZX6) Sentrin-specific protease 2 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP2)
(SUMO-1/Smt3-specific isopeptidase 2) (Smt3ip2) (Axam2)
Length = 588
Score = 48.9 bits (115), Expect = 3e-05
Identities = 36/118 (30%), Positives = 59/118 (49%), Gaps = 9/118 (7%)
Query: 223 GRLKFEIQLNEKRKSAFDLLRPKKELVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVN 282
G L+ +I + E ++ F P KE ++ E T++ E E++ A +++
Sbjct: 336 GLLRRKISVLEIKEKNF----PSKE--KDRRTEDLFEFTEDMEKEISNALGHGPPDEILS 389
Query: 283 HEKSNIEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
+ I+ + L+ WLNDEVIN Y+ LL ER ++ Q + H F+TFFY K
Sbjct: 390 -SAFKLRITRGDIQTLKNYHWLNDEVINFYMNLLVERSKK--QGYPALHAFSTFFYPK 444
>SEN2_RAT (Q9EQE1) Sentrin-specific protease 2 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP2) (Axin associating
molecule) (Axam)
Length = 588
Score = 48.9 bits (115), Expect = 3e-05
Identities = 36/118 (30%), Positives = 59/118 (49%), Gaps = 9/118 (7%)
Query: 223 GRLKFEIQLNEKRKSAFDLLRPKKELVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVN 282
G L+ +I + E ++ F P KE ++ E LT++ E E++ A +++
Sbjct: 336 GLLRRKISVLEAKEKNF----PSKE--KDRRTEDLFELTEDMEKEISNALGHGPPDEILS 389
Query: 283 HEKSNIEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
+ I+ + L+ WLNDEVIN Y+ LL ER ++ Q + H +TFFY K
Sbjct: 390 -SAFKLRITRGDIQTLKNYHWLNDEVINFYMNLLVERSKK--QGYPALHALSTFFYPK 444
>SEN6_HUMAN (Q9GZR1) Sentrin-specific protease 6 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP6) (SUMO-1 specific
protease 1) (Protease FKSG6)
Length = 1112
Score = 43.1 bits (100), Expect = 0.001
Identities = 21/57 (36%), Positives = 36/57 (62%), Gaps = 3/57 (5%)
Query: 285 KSNIEISGKMFRCLRPGEWLNDEVINLYLE-LLKERERREPQKFLKCHFFNTFFYKK 340
K I ++ + CL GE+LND +I+ YL+ L+ E+ ++E + H F++FFYK+
Sbjct: 663 KGGISVTNEDLHCLNEGEFLNDVIIDFYLKYLVLEKLKKEDAD--RIHIFSSFFYKR 717
>SEN7_HUMAN (Q9BQF6) Sentrin-specific protease 7 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP7) (SUMO-1 specific
protease 2)
Length = 984
Score = 40.4 bits (93), Expect = 0.010
Identities = 19/55 (34%), Positives = 32/55 (57%), Gaps = 1/55 (1%)
Query: 285 KSNIEISGKMFRCLRPGEWLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYK 339
K + ++ + CL GE+LND +I+ YL+ L E+ + + H F++FFYK
Sbjct: 691 KGGLGVTNEDLECLEEGEFLNDVIIDFYLKYL-ILEKASDELVERSHIFSSFFYK 744
>GCC2_MOUSE (Q8CHG3) GRIP and coiled-coil domain-containing protein 2
(Golgi coiled coil protein GCC185)
Length = 1679
Score = 40.0 bits (92), Expect = 0.013
Identities = 53/242 (21%), Positives = 106/242 (42%), Gaps = 31/242 (12%)
Query: 106 EKYEHAKHAALATISFREKGKVKDNIKEVVVLKDS--NVEDVEEFLSVVK---------- 153
EK +H L + + + +++D E+++LKDS V++ LS+VK
Sbjct: 889 EKVLVIEHENLRLLLKQRESELQDVRAELILLKDSLEKSPSVKDQLSLVKELEEKIESLE 948
Query: 154 ------DHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKM-LDNLSLSTKNDLSGVC 206
D K+ + LV + K E+ + + Q+ +++ + D LS S K L G
Sbjct: 949 KESKDKDEKISKIKLVAVKAKKELDSNRKEAQTLREELESVRSEKDRLSASMKEFLQGAE 1008
Query: 207 AYKKLLEAVDKRGDTLGRLKFEIQLNEKRKSAFDLLRPKKELVEEVPRE--PFVPLTKEE 264
+YK LL DK+ + QL+ +++ A + R ++L +++ + LT +
Sbjct: 1009 SYKSLLLEYDKQSE---------QLDVEKERAHNFERHIEDLTKQLRNSTCQYERLTSDN 1059
Query: 265 EVEVTRAFSANQNKVLVNHEKSNIE-ISGKMFRCLRPGEWLNDEVINLYLELLKERERRE 323
E + R + N L+ + ++ G + + L E ++ I ++ + E E +
Sbjct: 1060 EDLLARIETLQANAKLLEAQILEVQKAKGVVEKELDAEELQKEQKIKEHVSTVNELEELQ 1119
Query: 324 PQ 325
Q
Sbjct: 1120 LQ 1121
>SCP1_MOUSE (Q62209) Synaptonemal complex protein 1 (SCP-1 protein)
Length = 993
Score = 38.5 bits (88), Expect = 0.036
Identities = 47/222 (21%), Positives = 97/222 (43%), Gaps = 20/222 (9%)
Query: 80 KFDLSTNLKLTESSNLMASDT--------TDELMEKYEHAKHAALATISFREKGKVKDNI 131
KF + ++L E N++A D ++L E+ + + + REK +V D
Sbjct: 421 KFKNNKEVELEELKNILAEDQKLLDEKKQVEKLAEELQEKEQELTFLLETREK-EVHDLQ 479
Query: 132 KEVVVLKDSN---VEDVEEFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAA 188
++V V K S ++ VEE + ++ KLK L D + + N Q+++ ++
Sbjct: 480 EQVTVTKTSEQHYLKQVEEMKTELEKEKLKNTELTASCDMLLLENKKFVQEASDMALELK 539
Query: 189 KMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFEIQ-LNEKRKSAFDLLRPKKE 247
K +++ K + ++LL+ ++ + L+ E++ + ++ D ++ K +
Sbjct: 540 KHQEDIINCKKQE-------ERLLKQIENLEEKEMHLRDELESVRKEFIQQGDEVKCKLD 592
Query: 248 LVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSNIE 289
EE R + K+E+ N K V ++ NIE
Sbjct: 593 KSEENARSIECEVLKKEKQMKILESKCNNLKKQVENKSKNIE 634
>YJ9C_YEAST (P47166) Hypothetical 81.2 kDa protein in NMD5-HOM6
intergenic region
Length = 707
Score = 38.1 bits (87), Expect = 0.048
Identities = 64/279 (22%), Positives = 106/279 (37%), Gaps = 40/279 (14%)
Query: 44 PPPLTTNATVFRISRYPKTKPPLRREIHAPCSSRPRKFDLSTNLKLTESSNLMASDTTDE 103
P L N T F + K P ++ + + + ++ T+SS D+
Sbjct: 110 PTWLPKNYTEFTVEELVKEISPEYLRLNKQIDDLTNELNRKSQIETTDSSFFKLIKEKDD 169
Query: 104 LMEKYEHAKHAALATISFREKG-------KVKDNIKEVVVLKDSNVEDVEEFLSVVKDHK 156
L+++ + A LA R+ KVKD EV L DS+ + VE +
Sbjct: 170 LIDQLRK-EGAKLAETELRQSNQIKALRTKVKDLEYEVSELNDSSAQSVENY-------- 220
Query: 157 LKENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVD 216
N L L ++ G ++T+ + DA K ++L KN K+ + +
Sbjct: 221 ---NELQSLYHNIQ----GQLAEATNKLKDADKQKESLETLEKN-------IKEKDDLIT 266
Query: 217 KRGDTLGRLKFEIQLNEKRKSAFDLLRPKKELVEEVPREPFVPLTKEEEVEV-----TRA 271
+L ++ L EK KS F KK L E + TK E++ + T+
Sbjct: 267 ILQQSLDNMR---TLLEKEKSEFQ--TEKKALQEATVDQVTTLETKLEQLRIELDSSTQN 321
Query: 272 FSANQNKVLVNHEKSNIEISGKMFRCLRPGEWLNDEVIN 310
A N+ V+ ++S E F+ R E L N
Sbjct: 322 LDAKSNRDFVDDQQSYEEKQHASFQYNRLKEQLESSKAN 360
>SCP1_MESAU (Q60563) Synaptonemal complex protein 1 (SCP-1 protein)
(Meiotic chromosome synaptic protein) (Fragment)
Length = 845
Score = 38.1 bits (87), Expect = 0.048
Identities = 49/219 (22%), Positives = 93/219 (42%), Gaps = 21/219 (9%)
Query: 80 KFDLSTNLKLTESSNLMASDT--------TDELMEKYEHAKHAALATISFREKGKVKDNI 131
KF + +KL E ++A D ++L E+ + + + REK +V D
Sbjct: 277 KFKNNNEVKLEELKKILAEDQKLLDEKKQVEKLAEELQGKEQELTLLLQTREK-EVHDLE 335
Query: 132 KEVVVLKDSNV---EDVEEFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAA 188
++++V K S+ + VEE + +++ KLK L K+ + N + Q++ ++
Sbjct: 336 EQLLVTKISDQNYSKQVEELKTKLEEEKLKNAELTASCGKLSLENNKLTQETNDMALELK 395
Query: 189 KMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFE-IQLNEKRKSAFDLLRPKKE 247
K ++++ S K + + + L E D L ++ E IQ + K D
Sbjct: 396 KYQEDITNSKKQEERMLKQIENLEEKETHLRDELESVRKEFIQQGNEVKCKLDKSEENAR 455
Query: 248 LVE-EVPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEK 285
+E EV L KE+++++ N K N K
Sbjct: 456 SIECEV-------LKKEKQMKILENKCNNLRKQAENKSK 487
>SEN5_MACFA (Q8WP32) Sentrin-specific protease 5 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP5) (QtsA-16408)
Length = 755
Score = 37.4 bits (85), Expect = 0.081
Identities = 51/191 (26%), Positives = 75/191 (38%), Gaps = 54/191 (28%)
Query: 159 ENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVDKR 218
E LVC K+E GG + +S VD ++ VC L E + K
Sbjct: 468 EAPLVCSGLKLENQVGGGKDSQKASPVDDEQL-------------SVCLSGFLDEVMKKY 514
Query: 219 GDTLGRLKFEIQLNEKRKSAFDLLRPKKELVEE--VPREPFVPLTKEEEVEVTRAFSANQ 276
G + L+EK ++L K++ E R+PF+ E+ RA
Sbjct: 515 GSL-------VPLSEK-----EVLGRLKDVFNEDFSNRKPFI----NREITNYRA----- 553
Query: 277 NKVLVNHEKSNIEISGKM-------FRCLRPGEWLNDEVINLYLELLKERERREPQKFLK 329
H+K N I L WLND+VIN+Y EL+ + K
Sbjct: 554 -----RHQKCNFRIFYNKHMLDMDDLATLDGQNWLNDQVINMYGELIMDAVPD------K 602
Query: 330 CHFFNTFFYKK 340
HFFN+FF+++
Sbjct: 603 VHFFNSFFHRQ 613
>SEN5_HUMAN (Q96HI0) Sentrin-specific protease 5 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP5) (Protease FKSG45)
Length = 755
Score = 37.4 bits (85), Expect = 0.081
Identities = 51/191 (26%), Positives = 75/191 (38%), Gaps = 54/191 (28%)
Query: 159 ENGLVCLDDKVEVVNGGIQQQSTSSMVDAAKMLDNLSLSTKNDLSGVCAYKKLLEAVDKR 218
E LVC K+E GG + +S VD ++ VC L E + K
Sbjct: 468 EAPLVCSGLKLENQVGGGKNSQKASPVDDEQL-------------SVCLSGFLDEVMKKY 514
Query: 219 GDTLGRLKFEIQLNEKRKSAFDLLRPKKELVEE--VPREPFVPLTKEEEVEVTRAFSANQ 276
G + L+EK ++L K++ E R+PF+ E+ RA
Sbjct: 515 GSL-------VPLSEK-----EVLGRLKDVFNEDFSNRKPFI----NREITNYRA----- 553
Query: 277 NKVLVNHEKSNIEISGKM-------FRCLRPGEWLNDEVINLYLELLKERERREPQKFLK 329
H+K N I L WLND+VIN+Y EL+ + K
Sbjct: 554 -----RHQKCNFRIFYNKHMLDMDDLATLDGQNWLNDQVINMYGELIMDAVPD------K 602
Query: 330 CHFFNTFFYKK 340
HFFN+FF+++
Sbjct: 603 VHFFNSFFHRQ 613
>SEN3_MOUSE (Q9EP97) Sentrin-specific protease 3 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP3) (SUMO-1 specific
protease 3) (Smt3-specific isopeptidase 1) (Smt3ip1)
Length = 568
Score = 37.4 bits (85), Expect = 0.081
Identities = 17/38 (44%), Positives = 24/38 (62%), Gaps = 6/38 (15%)
Query: 303 WLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
WLND+V+N+Y +L+ + K HFFN+FFY K
Sbjct: 395 WLNDQVMNMYGDLVMDTVPE------KVHFFNSFFYDK 426
>SEN3_HUMAN (Q9H4L4) Sentrin-specific protease 3 (EC 3.4.22.-)
(Sentrin/SUMO-specific protease SENP3) (SUMO-1 specific
protease 3)
Length = 574
Score = 37.4 bits (85), Expect = 0.081
Identities = 17/38 (44%), Positives = 24/38 (62%), Gaps = 6/38 (15%)
Query: 303 WLNDEVINLYLELLKERERREPQKFLKCHFFNTFFYKK 340
WLND+V+N+Y +L+ + K HFFN+FFY K
Sbjct: 401 WLNDQVMNMYGDLVMDTVPE------KVHFFNSFFYDK 432
>ULP2_SCHPO (O13769) Ubiquitin-like-specific protease 2 (EC
3.4.22.-)
Length = 652
Score = 37.0 bits (84), Expect = 0.11
Identities = 19/56 (33%), Positives = 30/56 (52%), Gaps = 1/56 (1%)
Query: 286 SNIEISGKMFRCLRPGEWLNDEVINLYLELLK-ERERREPQKFLKCHFFNTFFYKK 340
++I I+ L GE+LND +++ YL L + + + P H FNTFFY +
Sbjct: 359 NSIAITNTDLTRLNEGEFLNDTIVDFYLRYLYCKLQTQNPSLANDTHIFNTFFYNR 414
>SCP1_RAT (Q03410) Synaptonemal complex protein 1 (SCP-1 protein)
Length = 997
Score = 36.6 bits (83), Expect = 0.14
Identities = 51/245 (20%), Positives = 103/245 (41%), Gaps = 20/245 (8%)
Query: 80 KFDLSTNLKLTESSNLMASDT--------TDELMEKYEHAKHAALATISFREKGKVKDNI 131
KF + ++L E ++A D ++L E+ + + + REK ++ D
Sbjct: 425 KFKNNKEVELEELKTILAEDQKLLDEKKQVEKLAEELQGKEQELTFLLQTREK-EIHDLE 483
Query: 132 KEVVVLKDSN---VEDVEEFLSVVKDHKLKENGLVCLDDKVEVVNGGIQQQSTSSMVDAA 188
+V V K S ++ VEE + ++ KLK L D + + N + Q+++ +++
Sbjct: 484 VQVTVTKTSEEHYLKQVEEMKTELEKEKLKNIELTANSDMLLLENKKLVQEASDMVLELK 543
Query: 189 KMLDNLSLSTKNDLSGVCAYKKLLEAVDKRGDTLGRLKFE-IQLNEKRKSAFDLLRPKKE 247
K +++ K + + + L E D L ++ E IQ ++ K D
Sbjct: 544 KHQEDIINCKKQEERMLKQIETLEEKEMNLRDELESVRKEFIQQGDEVKCKLDKSEENAR 603
Query: 248 LVEEVPREPFVPLTKEEEVEVTRAFSANQNKVLVNHEKSNIEISGKMFRCLRPGEWLNDE 307
+E + L KE+++++ N K + N K NIE + + L+ ++
Sbjct: 604 SIE------YEVLKKEKQMKILENKCNNLKKQIENKSK-NIEELHQENKALKKKSSAENK 656
Query: 308 VINLY 312
+N Y
Sbjct: 657 QLNAY 661
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,567,405
Number of Sequences: 164201
Number of extensions: 2187237
Number of successful extensions: 9198
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 82
Number of HSP's that attempted gapping in prelim test: 9139
Number of HSP's gapped (non-prelim): 124
length of query: 459
length of database: 59,974,054
effective HSP length: 114
effective length of query: 345
effective length of database: 41,255,140
effective search space: 14233023300
effective search space used: 14233023300
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC147012.13


