

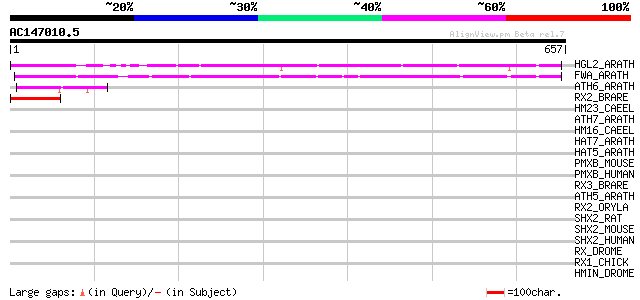
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.5 - phase: 0
(657 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
HGL2_ARATH (P46607) Homeobox protein GLABRA2 (Homeobox-leucine z... 322 2e-87
FWA_ARATH (Q9FVI6) Homeobox protein FWA 269 2e-71
ATH6_ARATH (P46668) Homeobox-leucine zipper protein ATHB-6 (Home... 47 2e-04
RX2_BRARE (O42357) Retinal homeobox protein Rx2 45 8e-04
HM23_CAEEL (P34663) Homeobox protein ceh-23 42 0.004
ATH7_ARATH (P46897) Homeobox-leucine zipper protein ATHB-7 (Home... 42 0.005
HM16_CAEEL (P34326) Homeobox protein engrailed-like ceh-16 41 0.011
HAT7_ARATH (Q00466) Homeobox-leucine zipper protein HAT7 (HD-ZIP... 41 0.011
HAT5_ARATH (Q02283) Homeobox-leucine zipper protein HAT5 (HD-ZIP... 41 0.011
PMXB_MOUSE (O35690) Paired mesoderm homeobox protein 2B (Paired-... 40 0.019
PMXB_HUMAN (Q99453) Paired mesoderm homeobox protein 2B (Paired-... 40 0.019
RX3_BRARE (O42358) Retinal homeobox protein Rx3 40 0.025
ATH5_ARATH (P46667) Homeobox-leucine zipper protein ATHB-5 (HD-Z... 40 0.025
RX2_ORYLA (Q9I9A2) Retinal homeobox protein Rx2 39 0.033
SHX2_RAT (O35750) Short stature homeobox protein 2 (Paired famil... 39 0.043
SHX2_MOUSE (P70390) Short stature homeobox protein 2 (Homeobox p... 39 0.043
SHX2_HUMAN (O60902) Short stature homeobox protein 2 (Paired-rel... 39 0.043
RX_DROME (Q9W2Q1) Retinal homeobox protein Rx (DRx1) (DRx) 39 0.043
RX1_CHICK (Q9PVY0) Retinal homeobox protein Rx1 (cRax1) 39 0.043
HMIN_DROME (P05527) Homeobox protein invected 39 0.043
>HGL2_ARATH (P46607) Homeobox protein GLABRA2 (Homeobox-leucine
zipper protein ATHB-10) (HD-ZIP protein ATHB-10)
Length = 745
Score = 322 bits (825), Expect = 2e-87
Identities = 221/664 (33%), Positives = 343/664 (51%), Gaps = 54/664 (8%)
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNENL 61
KE HPDE +R QL+ ++GL P+Q+KFWFQN+RT +K ER N L+ E +K+R EN
Sbjct: 119 KETPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKAELEKLREENK 178
Query: 62 KIKEVLKA--KICLDCGGPPFPMKDHQNFVQDLKQENAQLKQECEKMSSLLASYMEKKIS 119
++E C +CGG P DL EN++LK E +K+ + L
Sbjct: 179 AMRESFSKANSSCPNCGGGP----------DDLHLENSKLKAELDKLRAALG-------R 221
Query: 120 RPEFEQALKSIKSFSRDYECSSHVHGNLATWGGVLGQTSTQNYDAQKITMSQVVDAAMDE 179
P QA S S D E H G+L + GV + +K ++++ + A E
Sbjct: 222 TPYPLQA-----SCSDDQE---HRLGSLDFYTGV--------FALEKSRIAEISNRATLE 265
Query: 180 LVRLVRVNEPFWVKSPNTQDGYTFHRESYEQVFP--KNNHFKGANVCEESSKYSGLVKIS 237
L ++ EP W++S T + + Y + FP + + F G E+S+ +G+V +
Sbjct: 266 LQKMATSGEPMWLRSVET-GREILNYDEYLKEFPQAQASSFPGRKTI-EASRDAGIVFMD 323
Query: 238 GIDLVGMFLDSVKWTNLFPTIVTKAETIKVFEIGSPGSR-DGALLLMNEEMHILSPLVRP 296
L F+D +W F +++KA T+ V G SR DGA+ LM EM +L+P+V
Sbjct: 324 AHKLAQSFMDVGQWKETFACLISKAATVDVIRQGEGPSRIDGAIQLMFGEMQLLTPVVPT 383
Query: 297 REFNIIRYCKKFDAGVWVIADVSF---DSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVT 353
RE +R C++ W I DVS DS+ A L + K PSGCII + +G VT
Sbjct: 384 REVYFVRSCRQLSPEKWAIVDVSVSVEDSNTEKEASLLKCRKLPSGCIIEDTSNGHSKVT 443
Query: 354 WVEHVEVEDKIHTHYVYRDLVGNYNLYGAESWIKELQRMCERSLGSYVEAIPVEETIGVI 413
WVEH++V ++R LV +GA W+ LQ CER + +P ++++GV
Sbjct: 444 WVEHLDVSAST-VQPLFRSLVNTGLAFGARHWVATLQLHCERLVFFMATNVPTKDSLGV- 501
Query: 414 QTLEGRNSVIKLAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQPNG 473
TL GR SV+K+AQRM + F ++ + +T + +RVS R D +P G
Sbjct: 502 TTLAGRKSVLKMAQRMTQSFYRAIAASSYHQWTKITTKTGQDMRVSSRKNL-HDPGEPTG 560
Query: 474 TIVTAATTLWLPLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCISIIK 533
IV A+++LWLP+ +F+F +D +R +WD LS G + IA++S G GN ++ I+
Sbjct: 561 VIVCASSSLWLPVSPALLFDFFRDEARRHEWDALSNGAHVQSIANLSKGQDRGNSVA-IQ 619
Query: 534 PFIPTQRQMMILQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVCS--- 590
++ + +LQ+S T+ S ++YAP D T + L G D + ILP GF +
Sbjct: 620 TVKSREKSIWVLQDSSTNSYESVVVYAPVDINTTQLVLAGHDPSNIQILPSGFSIIPDGV 679
Query: 591 KSQPNLNAPFGASNNIEDGSLLTLAAQ-ILSTSPHEIDQVLNVEDITDINTHLATTILNV 649
+S+P + N + GSLLTLA Q +++ SP LN+E + + ++ T+ N+
Sbjct: 680 ESRPLVITSTQDDRNSQGGSLLTLALQTLINPSP---AAKLNMESVESVTNLVSVTLHNI 736
Query: 650 KDAL 653
K +L
Sbjct: 737 KRSL 740
>FWA_ARATH (Q9FVI6) Homeobox protein FWA
Length = 686
Score = 269 bits (688), Expect = 2e-71
Identities = 194/650 (29%), Positives = 314/650 (47%), Gaps = 33/650 (5%)
Query: 6 HPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNENLKIKE 65
HP E +R++L + + Q+K WFQNKR L K ++ N TLR E+D++ +++
Sbjct: 64 HPTEEQRYELGQRLNMGVNQVKNWFQNKRNLEKINNDHLENVTLREEHDRLLATQDQLRS 123
Query: 66 VLKAKICLDCGGPPFPMKDHQNFVQDLKQENAQLKQECEKMSSLLASYMEKKISRPEFEQ 125
+ +C CG D + VQ L ENA L++E ++ +S S+ ++++ EQ
Sbjct: 124 AMLRSLCNICGKAT-NCGDTEYEVQKLMAENANLEREIDQFNSRYLSHPKQRMVSTS-EQ 181
Query: 126 ALKSIKSFSRDYECSSHVHGNLATWGGVLGQTSTQNYDAQKITMSQVVDAAMDELVRLVR 185
A S SS+ N G T T + + A+ EL+ L
Sbjct: 182 APSS----------SSNPGINATPVLDFSGGTRTSEKETS--IFLNLAITALRELITLGE 229
Query: 186 VNEPFWVKSPNTQD-GYTFHRESYEQVFPKNNHFKGANVCEESSKYSGLVKISGIDLVGM 244
V+ PFW+ P + G + E Y F NN K E+S+ GLV ++ + LV
Sbjct: 230 VDCPFWMIDPIVRSKGVSKIYEKYRSSF--NNVTKPPGQIVEASRAKGLVPMTCVTLVKT 287
Query: 245 FLDSVKWTNLFPTIVTKAETIKVFEIGSPGSRDGALLLMNEEMHILSPLVRPREFNIIRY 304
+D+ KW N+F IV A T KV GS G++ G+L + E ++SPLV R+ IRY
Sbjct: 288 LMDTGKWVNVFAPIVPVASTHKVISTGSGGTKSGSLQQIQAEFQVISPLVPKRKVTFIRY 347
Query: 305 CKKFDAGVWVIADVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKI 364
CK+ G+WV+ DV+ + P P + PSG II ++ +G VTW+E E +
Sbjct: 348 CKEIRQGLWVVVDVT-PTQNPTLLPYGCSKRLPSGLIIDDLSNGYSQVTWIEQAEYNES- 405
Query: 365 HTHYVYRDLVGNYNLYGAESWIKELQRMCERSLGSYVEAIPVEETIGVIQTLEGRNSVIK 424
H H +Y+ L+G GA+ W+ LQR CE SL + E + G+ + +G ++K
Sbjct: 406 HIHQLYQPLIGYGIGLGAKRWLATLQRHCE-SLSTLSSTNLTEISPGL--SAKGATEIVK 462
Query: 425 LAQRMVKMFCESLTMPGQLELNHLTLASIGGIRVSFRSTTDDDTSQPNGTIVTAATTLWL 484
LAQRM + +T P + + + ++ ++ + G +++A+T++WL
Sbjct: 463 LAQRMTLNYYRGITSPSVDKWQKIQVENVAQNMSFMIRKNVNEPGELTGIVLSASTSVWL 522
Query: 485 PLPALKVFEFLKDPTKRSQWDGLSCGNPMHEIAHISNGPYHGNCISIIKPFIPTQRQMMI 544
P+ +F F+ + R +WD L+ M E I HGN IS++K M++
Sbjct: 523 PVNQHTLFAFISHLSFRHEWDILTNDTTMEETIRIQKAKRHGNIISLLK---IVNNGMLV 579
Query: 545 LQESFTSRVGSYIIYAPSDRQTMDVALRGEDSKELPILPYGFVVCSKSQPNLNAPFGASN 604
LQE + G+ ++YAP + ++++ RGE+S + LP GF + +N + N
Sbjct: 580 LQEIWNDASGAMVVYAPVETNSIELVKRGENSDSVKFLPSGFSIV---PDGVNGSYHRGN 636
Query: 605 NIEDGSLLTLAAQIL-STSPHEIDQVLNVEDITDINTHLATTILNVKDAL 653
G LLT QIL +P L + + T +A TI+ +K AL
Sbjct: 637 T-GGGCLLTFGLQILVGINP---TAALIQGTVKSVETLMAHTIVKIKSAL 682
>ATH6_ARATH (P46668) Homeobox-leucine zipper protein ATHB-6
(Homeodomain transcription factor ATHB-6) (HD-ZIP
protein ATHB-6)
Length = 311
Score = 46.6 bits (109), Expect = 2e-04
Identities = 35/122 (28%), Positives = 61/122 (49%), Gaps = 16/122 (13%)
Query: 9 EAERH-QLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIR---------N 58
E ER +LA E+GL+P+Q+ WFQN+R K + + G L+ + D +R N
Sbjct: 85 EPERKVKLAQELGLQPRQVAVWFQNRRARWKTKQLEKDYGVLKTQYDSLRHNFDSLRRDN 144
Query: 59 ENLKIKEVLKAKICLDCGGPPFPMKDHQNFVQ-----DLKQENAQLKQECEKMSSLLASY 113
E+L ++E+ K K L+ GG +++ V +K+E L ++ + S +
Sbjct: 145 ESL-LQEISKLKTKLNGGGGEEEEEENNAAVTTESDISVKEEEVSLPEKITEAPSSPPQF 203
Query: 114 ME 115
+E
Sbjct: 204 LE 205
>RX2_BRARE (O42357) Retinal homeobox protein Rx2
Length = 327
Score = 44.7 bits (104), Expect = 8e-04
Identities = 20/59 (33%), Positives = 37/59 (61%), Gaps = 1/59 (1%)
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNEN 60
++ H+PD R +LA++V L +++ WFQN+R + Q + +T GT++ + IR+ N
Sbjct: 155 EKSHYPDVYSREELAMKVNLPEVRVQVWFQNRRAKWRRQEKMDT-GTMKLHDSPIRSFN 212
>HM23_CAEEL (P34663) Homeobox protein ceh-23
Length = 305
Score = 42.4 bits (98), Expect = 0.004
Identities = 21/50 (42%), Positives = 32/50 (64%), Gaps = 5/50 (10%)
Query: 10 AERHQLAVEVGLEPKQIKFWFQNKRTLLKH---QHERETNGTLRRENDKI 56
AER LA +GL P Q++ WFQN+R+ KH Q E + + TL ++++I
Sbjct: 239 AERENLAQRLGLSPSQVRIWFQNRRS--KHRRKQQEEQQSTTLEEKSEEI 286
>ATH7_ARATH (P46897) Homeobox-leucine zipper protein ATHB-7
(Homeodomain transcription factor ATHB-7) (HD-ZIP
protein ATHB-7)
Length = 258
Score = 42.0 bits (97), Expect = 0.005
Identities = 20/58 (34%), Positives = 33/58 (56%)
Query: 14 QLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNENLKIKEVLKAKI 71
QLA E+GL+P+Q+ WFQNKR K + LR+ D + ++ +K+ +A +
Sbjct: 61 QLARELGLQPRQVAIWFQNKRARWKSKQLETEYNILRQNYDNLASQFESLKKEKQALV 118
>HM16_CAEEL (P34326) Homeobox protein engrailed-like ceh-16
Length = 203
Score = 40.8 bits (94), Expect = 0.011
Identities = 19/37 (51%), Positives = 23/37 (61%)
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLK 38
+E + E R +LA E+GL QIK WFQNKR LK
Sbjct: 123 RESRYLTEKRRQELAHELGLNESQIKIWFQNKRAKLK 159
>HAT7_ARATH (Q00466) Homeobox-leucine zipper protein HAT7 (HD-ZIP
protein 7) (HD-ZIP protein ATHB-3)
Length = 251
Score = 40.8 bits (94), Expect = 0.011
Identities = 20/53 (37%), Positives = 33/53 (61%), Gaps = 1/53 (1%)
Query: 9 EAERH-QLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKIRNEN 60
E ER QLA +GL+P+QI WFQN+R K + +L+++ D ++++N
Sbjct: 75 EPERKMQLAKALGLQPRQIAIWFQNRRARWKTKQLERDYDSLKKQFDVLKSDN 127
>HAT5_ARATH (Q02283) Homeobox-leucine zipper protein HAT5 (HD-ZIP
protein 5) (HD-ZIP protein ATHB-1)
Length = 272
Score = 40.8 bits (94), Expect = 0.011
Identities = 26/77 (33%), Positives = 42/77 (53%), Gaps = 15/77 (19%)
Query: 9 EAERH-QLAVEVGLEPKQIKFWFQNKRT------------LLKHQHER--ETNGTLRREN 53
E ER QLA ++GL+P+Q+ WFQN+R LLK +++ ++ +N
Sbjct: 91 EPERKTQLAKKLGLQPRQVAVWFQNRRARWKTKQLERDYDLLKSTYDQLLSNYDSIVMDN 150
Query: 54 DKIRNENLKIKEVLKAK 70
DK+R+E + E L+ K
Sbjct: 151 DKLRSEVTSLTEKLQGK 167
>PMXB_MOUSE (O35690) Paired mesoderm homeobox protein 2B
(Paired-like homeobox 2B) (PHOX2B homeodomain protein)
(Neuroblastoma Phox) (NBPhox)
Length = 314
Score = 40.0 bits (92), Expect = 0.019
Identities = 19/67 (28%), Positives = 35/67 (51%), Gaps = 7/67 (10%)
Query: 3 ECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERET-------NGTLRRENDK 55
E H+PD R +LA+++ L +++ WFQN+R + Q NG+ +++D
Sbjct: 119 ETHYPDIYTREELALKIDLTEARVQVWFQNRRAKFRKQERAAAAAAAAAKNGSSGKKSDS 178
Query: 56 IRNENLK 62
R++ K
Sbjct: 179 SRDDESK 185
>PMXB_HUMAN (Q99453) Paired mesoderm homeobox protein 2B
(Paired-like homeobox 2B) (PHOX2B homeodomain protein)
(Neuroblastoma Phox) (NBPhox)
Length = 314
Score = 40.0 bits (92), Expect = 0.019
Identities = 19/67 (28%), Positives = 35/67 (51%), Gaps = 7/67 (10%)
Query: 3 ECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERET-------NGTLRRENDK 55
E H+PD R +LA+++ L +++ WFQN+R + Q NG+ +++D
Sbjct: 119 ETHYPDIYTREELALKIDLTEARVQVWFQNRRAKFRKQERAAAAAAAAAKNGSSGKKSDS 178
Query: 56 IRNENLK 62
R++ K
Sbjct: 179 SRDDESK 185
>RX3_BRARE (O42358) Retinal homeobox protein Rx3
Length = 292
Score = 39.7 bits (91), Expect = 0.025
Identities = 16/55 (29%), Positives = 32/55 (58%)
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTLRRENDKI 56
++ H+PD R +LA++V L +++ WFQN+R + Q + E + +E+ +
Sbjct: 126 EKSHYPDVYSREELALKVNLPEVRVQVWFQNRRAKWRRQEKLEVSSIKLQESSML 180
>ATH5_ARATH (P46667) Homeobox-leucine zipper protein ATHB-5 (HD-ZIP
protein ATHB-5)
Length = 312
Score = 39.7 bits (91), Expect = 0.025
Identities = 30/75 (40%), Positives = 43/75 (57%), Gaps = 13/75 (17%)
Query: 9 EAERH-QLAVEVGLEPKQIKFWFQNKRTLLK-HQHERETN------GTLRRENDKIRNEN 60
E ER +LA E+GL+P+Q+ WFQN+R K Q ER+ L+R D ++ +N
Sbjct: 95 EPERKVKLAQELGLQPRQVAIWFQNRRARWKTKQLERDYGVLKSNFDALKRNRDSLQRDN 154
Query: 61 ----LKIKEVLKAKI 71
+IKE LKAK+
Sbjct: 155 DSLLGQIKE-LKAKL 168
>RX2_ORYLA (Q9I9A2) Retinal homeobox protein Rx2
Length = 327
Score = 39.3 bits (90), Expect = 0.033
Identities = 15/45 (33%), Positives = 28/45 (61%)
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETN 46
++ H+PD R +LA +V L +++ WFQN+R + Q + +T+
Sbjct: 157 EKSHYPDVYSREELATKVNLPEVRVQVWFQNRRAKWRRQEKMDTS 201
>SHX2_RAT (O35750) Short stature homeobox protein 2 (Paired family
homeodomain protein Prx3) (Fragment)
Length = 237
Score = 38.9 bits (89), Expect = 0.043
Identities = 16/47 (34%), Positives = 26/47 (55%)
Query: 3 ECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTL 49
E H+PD R +L+ +GL +++ WFQN+R + Q + G L
Sbjct: 67 ETHYPDAFMREELSQRLGLSEARVQVWFQNRRAKCRKQENQLHKGVL 113
>SHX2_MOUSE (P70390) Short stature homeobox protein 2 (Homeobox
protein Og12X) (OG-12) (Paired family homeodomain
protein Prx3)
Length = 331
Score = 38.9 bits (89), Expect = 0.043
Identities = 16/47 (34%), Positives = 26/47 (55%)
Query: 3 ECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTL 49
E H+PD R +L+ +GL +++ WFQN+R + Q + G L
Sbjct: 161 ETHYPDAFMREELSQRLGLSEARVQVWFQNRRAKCRKQENQLHKGVL 207
>SHX2_HUMAN (O60902) Short stature homeobox protein 2
(Paired-related homeobox protein SHOT) (Homeobox protein
Og12X)
Length = 331
Score = 38.9 bits (89), Expect = 0.043
Identities = 16/47 (34%), Positives = 26/47 (55%)
Query: 3 ECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETNGTL 49
E H+PD R +L+ +GL +++ WFQN+R + Q + G L
Sbjct: 161 ETHYPDAFMREELSQRLGLSEARVQVWFQNRRAKCRKQENQLHKGVL 207
>RX_DROME (Q9W2Q1) Retinal homeobox protein Rx (DRx1) (DRx)
Length = 873
Score = 38.9 bits (89), Expect = 0.043
Identities = 15/44 (34%), Positives = 28/44 (63%)
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERET 45
++ H+PD R +LA++V L +++ WFQN+R + Q + E+
Sbjct: 548 EKSHYPDVYSREELAMKVNLPEVRVQVWFQNRRAKWRRQEKSES 591
>RX1_CHICK (Q9PVY0) Retinal homeobox protein Rx1 (cRax1)
Length = 228
Score = 38.9 bits (89), Expect = 0.043
Identities = 15/45 (33%), Positives = 28/45 (61%)
Query: 2 KECHHPDEAERHQLAVEVGLEPKQIKFWFQNKRTLLKHQHERETN 46
++ H+PD R +LA++V L +++ WFQN+R + Q + E +
Sbjct: 56 EKSHYPDVYSREELAMKVNLPEVRVQVWFQNRRAKWRRQEKMEAS 100
>HMIN_DROME (P05527) Homeobox protein invected
Length = 576
Score = 38.9 bits (89), Expect = 0.043
Identities = 18/30 (60%), Positives = 20/30 (66%)
Query: 9 EAERHQLAVEVGLEPKQIKFWFQNKRTLLK 38
E R QL+ E+GL QIK WFQNKR LK
Sbjct: 498 EKRRQQLSGELGLNEAQIKIWFQNKRAKLK 527
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 79,328,040
Number of Sequences: 164201
Number of extensions: 3405737
Number of successful extensions: 9776
Number of sequences better than 10.0: 486
Number of HSP's better than 10.0 without gapping: 427
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 9304
Number of HSP's gapped (non-prelim): 515
length of query: 657
length of database: 59,974,054
effective HSP length: 117
effective length of query: 540
effective length of database: 40,762,537
effective search space: 22011769980
effective search space used: 22011769980
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147010.5


