

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147010.3 - phase: 0 /pseudo
(675 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
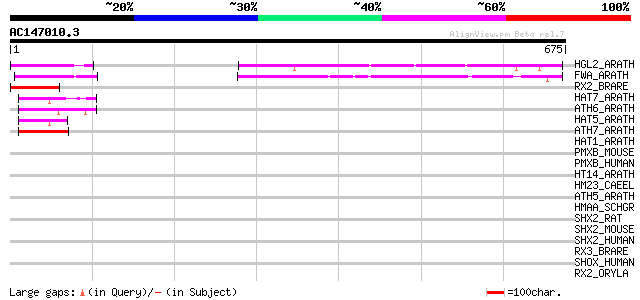
Score E
Sequences producing significant alignments: (bits) Value
HGL2_ARATH (P46607) Homeobox protein GLABRA2 (Homeobox-leucine z... 218 4e-56
FWA_ARATH (Q9FVI6) Homeobox protein FWA 180 9e-45
RX2_BRARE (O42357) Retinal homeobox protein Rx2 46 4e-04
HAT7_ARATH (Q00466) Homeobox-leucine zipper protein HAT7 (HD-ZIP... 46 4e-04
ATH6_ARATH (P46668) Homeobox-leucine zipper protein ATHB-6 (Home... 46 4e-04
HAT5_ARATH (Q02283) Homeobox-leucine zipper protein HAT5 (HD-ZIP... 45 5e-04
ATH7_ARATH (P46897) Homeobox-leucine zipper protein ATHB-7 (Home... 45 5e-04
HAT1_ARATH (P46600) Homeobox-leucine zipper protein HAT1 (HD-ZIP... 42 0.004
PMXB_MOUSE (O35690) Paired mesoderm homeobox protein 2B (Paired-... 42 0.005
PMXB_HUMAN (Q99453) Paired mesoderm homeobox protein 2B (Paired-... 42 0.005
HT14_ARATH (P46665) Homeobox-leucine zipper protein HAT14 (HD-ZI... 42 0.007
HM23_CAEEL (P34663) Homeobox protein ceh-23 42 0.007
ATH5_ARATH (P46667) Homeobox-leucine zipper protein ATHB-5 (HD-Z... 42 0.007
HMAA_SCHGR (P29556) Homeobox protein abdominal-A homolog (Fragment) 41 0.009
SHX2_RAT (O35750) Short stature homeobox protein 2 (Paired famil... 41 0.012
SHX2_MOUSE (P70390) Short stature homeobox protein 2 (Homeobox p... 41 0.012
SHX2_HUMAN (O60902) Short stature homeobox protein 2 (Paired-rel... 41 0.012
RX3_BRARE (O42358) Retinal homeobox protein Rx3 41 0.012
SHOX_HUMAN (O15266) Short stature homeobox protein (Short statur... 40 0.015
RX2_ORYLA (Q9I9A2) Retinal homeobox protein Rx2 40 0.015
>HGL2_ARATH (P46607) Homeobox protein GLABRA2 (Homeobox-leucine
zipper protein ATHB-10) (HD-ZIP protein ATHB-10)
Length = 745
Score = 218 bits (555), Expect = 4e-56
Identities = 135/404 (33%), Positives = 220/404 (54%), Gaps = 14/404 (3%)
Query: 279 FPTIVTKAETIKVFEIGSRGSR-DGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
F +++KA T+ V G SR DGA+ LM EM +L+P+V RE +R C+++ P W
Sbjct: 341 FACLISKAATVDVIRQGEGPSRIDGAIQLMFGEMQLLTPVVPTREVYFVRSCRQLSPEKW 400
Query: 338 VITDVSF---DSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVY 394
I DVS DS+ A L + K PSGCII + +G VTWVEH++V ++
Sbjct: 401 AIVDVSVSVEDSNTEKEASLLKCRKLPSGCIIEDTSNGHSKVTWVEHLDVSAST-VQPLF 459
Query: 395 RDLVGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMV 454
R LV +GA W+ LQ CER + +P ++++GV TL GR SV+K+A RM
Sbjct: 460 RSLVNTGLAFGARHWVATLQLHCERLVFFMATNVPTKDSLGVT-TLAGRKSVLKMAQRMT 518
Query: 455 KMFCECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQK 514
+ F + + +T + +RVS R D +P G +V A+++LWLP+
Sbjct: 519 QSFYRAIAASSYHQWTKITTKTGQDMRVSSRKNLHDPG-EPTGVIVCASSSLWLPVSPAL 577
Query: 515 VFEFLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQRQMVILQESFT 574
+F+F +D +R +W+ LS G + IA++S G GN ++ I++ ++ + +LQ+S T
Sbjct: 578 LFDFFRDEARRHEWDALSNGAHVQSIANLSKGQDRGNSVA-IQTVKSREKSIWVLQDSST 636
Query: 575 SSVGSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCS---KNQANLNAPFGASKSIE 631
+S S V+YAP+D T + L G D + ILP G + +++ + ++ +
Sbjct: 637 NSYESVVVYAPVDINTTQLVLAGHDPSNIQILPSGFSIIPDGVESRPLVITSTQDDRNSQ 696
Query: 632 DGSLITLAAQTY---AIGQVLNVDSLNDINSQLASTILNVKDAL 672
GSL+TLA QT + LN++S+ + + ++ T+ N+K +L
Sbjct: 697 GGSLLTLALQTLINPSPAAKLNMESVESVTNLVSVTLHNIKRSL 740
Score = 78.6 bits (192), Expect = 5e-14
Identities = 41/103 (39%), Positives = 58/103 (55%), Gaps = 12/103 (11%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KE HPDE QR QL+ ++GL P+Q+K WFQN+R +K ER N L+ E +K+R EN
Sbjct: 119 KETPHPDEKQRQQLSKQLGLAPRQVKFWFQNRRTQIKAIQERHENSLLKAELEKLREENK 178
Query: 62 KIKEELKA--KICLDCGGSPFPMKDHQNFVQEMKQENAQLKQE 102
++E C +CGG P ++ EN++LK E
Sbjct: 179 AMRESFSKANSSCPNCGGGP----------DDLHLENSKLKAE 211
>FWA_ARATH (Q9FVI6) Homeobox protein FWA
Length = 686
Score = 180 bits (457), Expect = 9e-45
Identities = 114/403 (28%), Positives = 202/403 (49%), Gaps = 25/403 (6%)
Query: 278 LFPTIVTKAETIKVFEIGSRGSRDGALLLMNEEMHILSPLVRPREFNIIRYCKKVDPGVW 337
+F IV A T KV GS G++ G+L + E ++SPLV R+ IRYCK++ G+W
Sbjct: 297 VFAPIVPVASTHKVISTGSGGTKSGSLQQIQAEFQVISPLVPKRKVTFIRYCKEIRQGLW 356
Query: 338 VITDVSFDSSRPNTAPLSRGWKHPSGCIIREMPHGGCLVTWVEHVEVEDKIHTHYVYRDL 397
V+ DV+ + P P + PSG II ++ +G VTW+E E + H H +Y+ L
Sbjct: 357 VVVDVT-PTQNPTLLPYGCSKRLPSGLIIDDLSNGYSQVTWIEQAEYNES-HIHQLYQPL 414
Query: 398 VGEYNLYGAESWIKELQRMCERSLGSNVEAIPVEETIGVIQTLEGRNSVIKLADRMVKMF 457
+G GA+ W+ LQR CE SL + E + G+ + +G ++KLA RM +
Sbjct: 415 IGYGIGLGAKRWLATLQRHCE-SLSTLSSTNLTEISPGL--SAKGATEIVKLAQRMTLNY 471
Query: 458 CECLTMPGQVELNHLTLDSIGGVRVSIRATTDDDASQPNGTVVTAATTLWLPLPAQKVFE 517
+T P + + ++++ + ++ + G V++A+T++WLP+ +F
Sbjct: 472 YRGITSPSVDKWQKIQVENVAQNMSFMIRKNVNEPGELTGIVLSASTSVWLPVNQHTLFA 531
Query: 518 FLKDPTKRSQWNGLSCGNPMHEIAHISNGPYHGNCISVIKSFIPTQRQMVILQESFTSSV 577
F+ + R +W+ L+ M E I HGN IS++K M++LQE + +
Sbjct: 532 FISHLSFRHEWDILTNDTTMEETIRIQKAKRHGNIISLLKI---VNNGMLVLQEIWNDAS 588
Query: 578 GSYVIYAPIDRKTMDVALRGEDSKELPILPYGLIVCSKNQANLNAPFGASKSIEDGSLIT 637
G+ V+YAP++ ++++ RGE+S + LP G + P G + S G+
Sbjct: 589 GAMVVYAPVETNSIELVKRGENSDSVKFLPSGFSI---------VPDGVNGSYHRGNTGG 639
Query: 638 LAAQTYAIGQVLNVD--------SLNDINSQLASTILNVKDAL 672
T+ + ++ ++ ++ + + +A TI+ +K AL
Sbjct: 640 GCLLTFGLQILVGINPTAALIQGTVKSVETLMAHTIVKIKSAL 682
Score = 63.9 bits (154), Expect = 1e-09
Identities = 33/101 (32%), Positives = 57/101 (55%), Gaps = 1/101 (0%)
Query: 6 HPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENLKIKE 65
HP E QR +L ++ + Q+K+WFQNKR + K ++ N TLR E+D++ +++
Sbjct: 64 HPTEEQRYELGQRLNMGVNQVKNWFQNKRNLEKINNDHLENVTLREEHDRLLATQDQLRS 123
Query: 66 ELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQEASEF 106
+ +C CG + D + VQ++ ENA L++E +F
Sbjct: 124 AMLRSLCNICGKAT-NCGDTEYEVQKLMAENANLEREIDQF 163
>RX2_BRARE (O42357) Retinal homeobox protein Rx2
Length = 327
Score = 45.8 bits (107), Expect = 4e-04
Identities = 21/59 (35%), Positives = 38/59 (63%), Gaps = 1/59 (1%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNEN 60
++ H+PD R +LA+K+ L +++ WFQN+RA + Q + +T GT++ + IR+ N
Sbjct: 155 EKSHYPDVYSREELAMKVNLPEVRVQVWFQNRRAKWRRQEKMDT-GTMKLHDSPIRSFN 212
>HAT7_ARATH (Q00466) Homeobox-leucine zipper protein HAT7 (HD-ZIP
protein 7) (HD-ZIP protein ATHB-3)
Length = 251
Score = 45.8 bits (107), Expect = 4e-04
Identities = 33/102 (32%), Positives = 53/102 (51%), Gaps = 25/102 (24%)
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLK-HQHERETNG------TLRRENDKIRNENLKI 63
++ QLA +GL+P+QI WFQN+RA K Q ER+ + L+ +ND + N K+
Sbjct: 78 RKMQLAKALGLQPRQIAIWFQNRRARWKTKQLERDYDSLKKQFDVLKSDNDSLLAHNKKL 137
Query: 64 KEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQEASE 105
EL A +K H ++E+A++K+E +E
Sbjct: 138 HAELVA------------LKKHD------RKESAKIKREFAE 161
>ATH6_ARATH (P46668) Homeobox-leucine zipper protein ATHB-6
(Homeodomain transcription factor ATHB-6) (HD-ZIP
protein ATHB-6)
Length = 311
Score = 45.8 bits (107), Expect = 4e-04
Identities = 31/109 (28%), Positives = 57/109 (51%), Gaps = 15/109 (13%)
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIR---------NENL 61
++ +LA ++GL+P+Q+ WFQN+RA K + + G L+ + D +R NE+L
Sbjct: 88 RKVKLAQELGLQPRQVAVWFQNRRARWKTKQLEKDYGVLKTQYDSLRHNFDSLRRDNESL 147
Query: 62 KIKEELKAKICLDCGGSPFPMKDHQNFVQ-----EMKQENAQLKQEASE 105
++E K K L+ GG +++ V +K+E L ++ +E
Sbjct: 148 -LQEISKLKTKLNGGGGEEEEEENNAAVTTESDISVKEEEVSLPEKITE 195
>HAT5_ARATH (Q02283) Homeobox-leucine zipper protein HAT5 (HD-ZIP
protein 5) (HD-ZIP protein ATHB-1)
Length = 272
Score = 45.4 bits (106), Expect = 5e-04
Identities = 26/74 (35%), Positives = 43/74 (57%), Gaps = 14/74 (18%)
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLK-HQHERETN-------------GTLRRENDKI 56
++ QLA K+GL+P+Q+ WFQN+RA K Q ER+ + ++ +NDK+
Sbjct: 94 RKTQLAKKLGLQPRQVAVWFQNRRARWKTKQLERDYDLLKSTYDQLLSNYDSIVMDNDKL 153
Query: 57 RNENLKIKEELKAK 70
R+E + E+L+ K
Sbjct: 154 RSEVTSLTEKLQGK 167
>ATH7_ARATH (P46897) Homeobox-leucine zipper protein ATHB-7
(Homeodomain transcription factor ATHB-7) (HD-ZIP
protein ATHB-7)
Length = 258
Score = 45.4 bits (106), Expect = 5e-04
Identities = 21/61 (34%), Positives = 37/61 (60%)
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENLKIKEELKAK 70
++ QLA ++GL+P+Q+ WFQNKRA K + LR+ D + ++ +K+E +A
Sbjct: 58 KKVQLARELGLQPRQVAIWFQNKRARWKSKQLETEYNILRQNYDNLASQFESLKKEKQAL 117
Query: 71 I 71
+
Sbjct: 118 V 118
>HAT1_ARATH (P46600) Homeobox-leucine zipper protein HAT1 (HD-ZIP
protein 1)
Length = 282
Score = 42.4 bits (98), Expect = 0.004
Identities = 22/65 (33%), Positives = 38/65 (57%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENL 61
KE + + Q+ LA K+GL +Q++ WFQN+RA K + L+R +K+ EN
Sbjct: 152 KEHNTLNPKQKLALAKKLGLTARQVEVWFQNRRARTKLKQTEVDCEYLKRCVEKLTEENR 211
Query: 62 KIKEE 66
++++E
Sbjct: 212 RLEKE 216
>PMXB_MOUSE (O35690) Paired mesoderm homeobox protein 2B
(Paired-like homeobox 2B) (PHOX2B homeodomain protein)
(Neuroblastoma Phox) (NBPhox)
Length = 314
Score = 42.0 bits (97), Expect = 0.005
Identities = 22/67 (32%), Positives = 36/67 (52%), Gaps = 7/67 (10%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERET-------NGTLRRENDK 55
E H+PD R +LA+KI L +++ WFQN+RA + Q NG+ +++D
Sbjct: 119 ETHYPDIYTREELALKIDLTEARVQVWFQNRRAKFRKQERAAAAAAAAAKNGSSGKKSDS 178
Query: 56 IRNENLK 62
R++ K
Sbjct: 179 SRDDESK 185
>PMXB_HUMAN (Q99453) Paired mesoderm homeobox protein 2B
(Paired-like homeobox 2B) (PHOX2B homeodomain protein)
(Neuroblastoma Phox) (NBPhox)
Length = 314
Score = 42.0 bits (97), Expect = 0.005
Identities = 22/67 (32%), Positives = 36/67 (52%), Gaps = 7/67 (10%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERET-------NGTLRRENDK 55
E H+PD R +LA+KI L +++ WFQN+RA + Q NG+ +++D
Sbjct: 119 ETHYPDIYTREELALKIDLTEARVQVWFQNRRAKFRKQERAAAAAAAAAKNGSSGKKSDS 178
Query: 56 IRNENLK 62
R++ K
Sbjct: 179 SRDDESK 185
>HT14_ARATH (P46665) Homeobox-leucine zipper protein HAT14 (HD-ZIP
protein 14)
Length = 225
Score = 41.6 bits (96), Expect = 0.007
Identities = 19/58 (32%), Positives = 35/58 (59%)
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKIRNENLKIKEELK 68
Q+ LA ++ L P+Q++ WFQN+RA K + L+R + + EN ++++E+K
Sbjct: 105 QKIALAKQLNLRPRQVEVWFQNRRARTKLKQTEVDCEYLKRCCESLTEENRRLQKEVK 162
>HM23_CAEEL (P34663) Homeobox protein ceh-23
Length = 305
Score = 41.6 bits (96), Expect = 0.007
Identities = 20/58 (34%), Positives = 36/58 (61%), Gaps = 1/58 (1%)
Query: 10 AQRCQLAVKIGLEPKQIKSWFQNKRAM-LKHQHERETNGTLRRENDKIRNENLKIKEE 66
A+R LA ++GL P Q++ WFQN+R+ + Q E + + TL ++++I + + EE
Sbjct: 239 AERENLAQRLGLSPSQVRIWFQNRRSKHRRKQQEEQQSTTLEEKSEEIGKDEEEDDEE 296
>ATH5_ARATH (P46667) Homeobox-leucine zipper protein ATHB-5 (HD-ZIP
protein ATHB-5)
Length = 312
Score = 41.6 bits (96), Expect = 0.007
Identities = 26/71 (36%), Positives = 42/71 (58%), Gaps = 10/71 (14%)
Query: 11 QRCQLAVKIGLEPKQIKSWFQNKRAMLK-HQHERETN------GTLRRENDKIRNEN--- 60
++ +LA ++GL+P+Q+ WFQN+RA K Q ER+ L+R D ++ +N
Sbjct: 98 RKVKLAQELGLQPRQVAIWFQNRRARWKTKQLERDYGVLKSNFDALKRNRDSLQRDNDSL 157
Query: 61 LKIKEELKAKI 71
L +ELKAK+
Sbjct: 158 LGQIKELKAKL 168
>HMAA_SCHGR (P29556) Homeobox protein abdominal-A homolog (Fragment)
Length = 157
Score = 41.2 bits (95), Expect = 0.009
Identities = 29/101 (28%), Positives = 50/101 (48%), Gaps = 16/101 (15%)
Query: 5 HHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHE--RETNGTLRRENDKIRNENLK 62
H+ +R ++A + L +QIK WFQN+R LK + +E N RRE R E +
Sbjct: 29 HYLTRRRRIEIAHALCLTERQIKIWFQNRRMKLKKELRAVKEINEQARRE----REEQDR 84
Query: 63 IKEELKAKICLDCGGSPFPMKDHQNFVQEMKQENAQLKQEA 103
+K++ + K+ + Q Q+ +Q+ Q +Q+A
Sbjct: 85 LKQQQEKKL----------EQQQQQQQQQQQQQQQQQQQQA 115
>SHX2_RAT (O35750) Short stature homeobox protein 2 (Paired family
homeodomain protein Prx3) (Fragment)
Length = 237
Score = 40.8 bits (94), Expect = 0.012
Identities = 17/47 (36%), Positives = 28/47 (59%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTL 49
E H+PD R +L+ ++GL +++ WFQN+RA + Q + G L
Sbjct: 67 ETHYPDAFMREELSQRLGLSEARVQVWFQNRRAKCRKQENQLHKGVL 113
>SHX2_MOUSE (P70390) Short stature homeobox protein 2 (Homeobox
protein Og12X) (OG-12) (Paired family homeodomain
protein Prx3)
Length = 331
Score = 40.8 bits (94), Expect = 0.012
Identities = 17/47 (36%), Positives = 28/47 (59%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTL 49
E H+PD R +L+ ++GL +++ WFQN+RA + Q + G L
Sbjct: 161 ETHYPDAFMREELSQRLGLSEARVQVWFQNRRAKCRKQENQLHKGVL 207
>SHX2_HUMAN (O60902) Short stature homeobox protein 2
(Paired-related homeobox protein SHOT) (Homeobox protein
Og12X)
Length = 331
Score = 40.8 bits (94), Expect = 0.012
Identities = 17/47 (36%), Positives = 28/47 (59%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTL 49
E H+PD R +L+ ++GL +++ WFQN+RA + Q + G L
Sbjct: 161 ETHYPDAFMREELSQRLGLSEARVQVWFQNRRAKCRKQENQLHKGVL 207
>RX3_BRARE (O42358) Retinal homeobox protein Rx3
Length = 292
Score = 40.8 bits (94), Expect = 0.012
Identities = 17/55 (30%), Positives = 33/55 (59%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTLRRENDKI 56
++ H+PD R +LA+K+ L +++ WFQN+RA + Q + E + +E+ +
Sbjct: 126 EKSHYPDVYSREELALKVNLPEVRVQVWFQNRRAKWRRQEKLEVSSIKLQESSML 180
>SHOX_HUMAN (O15266) Short stature homeobox protein (Short stature
homeobox containing protein) (Pseudoautosomal homeobox
containing osteogenic protein)
Length = 292
Score = 40.4 bits (93), Expect = 0.015
Identities = 16/47 (34%), Positives = 28/47 (59%)
Query: 3 ECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETNGTL 49
E H+PD R +L+ ++GL +++ WFQN+RA + Q + G +
Sbjct: 138 ETHYPDAFMREELSQRLGLSEARVQVWFQNRRAKCRKQENQMHKGVI 184
>RX2_ORYLA (Q9I9A2) Retinal homeobox protein Rx2
Length = 327
Score = 40.4 bits (93), Expect = 0.015
Identities = 16/45 (35%), Positives = 29/45 (63%)
Query: 2 KECHHPDEAQRCQLAVKIGLEPKQIKSWFQNKRAMLKHQHERETN 46
++ H+PD R +LA K+ L +++ WFQN+RA + Q + +T+
Sbjct: 157 EKSHYPDVYSREELATKVNLPEVRVQVWFQNRRAKWRRQEKMDTS 201
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.330 0.142 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 76,821,726
Number of Sequences: 164201
Number of extensions: 3160698
Number of successful extensions: 10186
Number of sequences better than 10.0: 498
Number of HSP's better than 10.0 without gapping: 441
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 9704
Number of HSP's gapped (non-prelim): 522
length of query: 675
length of database: 59,974,054
effective HSP length: 117
effective length of query: 558
effective length of database: 40,762,537
effective search space: 22745495646
effective search space used: 22745495646
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147010.3


