

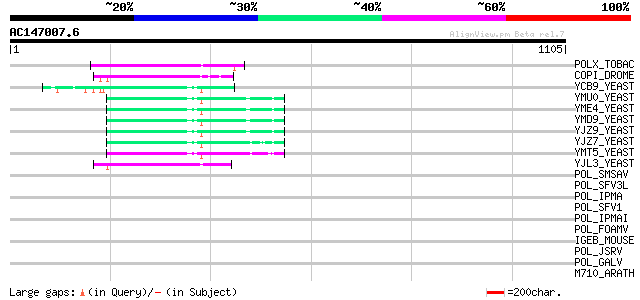
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147007.6 - phase: 0 /pseudo
(1105 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 213 2e-54
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 147 1e-34
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 92 1e-17
YMU0_YEAST (Q04670) Transposon Ty1 protein B 86 7e-16
YME4_YEAST (Q04711) Transposon Ty1 protein B 86 7e-16
YMD9_YEAST (Q03434) Transposon Ty1 protein B 86 7e-16
YJZ9_YEAST (P47100) Transposon Ty1 protein B 86 7e-16
YJZ7_YEAST (P47098) Transposon Ty1 protein B 85 9e-16
YMT5_YEAST (Q04214) Transposon Ty1 protein B 84 2e-15
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 62 1e-08
POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse transcript... 42 0.007
POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.2... 42 0.009
POL_IPMA (P11368) Putative Pol polyprotein [Contains: Integrase ... 42 0.009
POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23... 42 0.012
POL_IPMAI (P12894) Probable Pol polyprotein [Contains: Integrase... 42 0.012
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 42 0.012
IGEB_MOUSE (P03975) IgE-binding protein 42 0.012
POL_JSRV (P31623) Pol polyprotein [Contains: Reverse transcripta... 41 0.020
POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC 3.4.23... 41 0.020
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 40 0.027
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 213 bits (542), Expect = 2e-54
Identities = 125/327 (38%), Positives = 180/327 (54%), Gaps = 24/327 (7%)
Query: 162 SPDIIHCRLGHPSLDKLKVLVPH--LSHLKSLD---CESCQLGKHVRVSFPSRANKRSMS 216
S D+ H R+GH S L++L +S+ K C+ C GK RVSF + +++R ++
Sbjct: 421 SVDLWHKRMGHMSEKGLQILAKKSLISYAKGTTVKPCDYCLFGKQHRVSFQT-SSERKLN 479
Query: 217 PFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSRCTWITLLKDRS*LFGAFQTFCS*IKTQ 276
++++SDV GP + S G KY+VTFIDD SR W+ +LK + +F FQ F + ++ +
Sbjct: 480 ILDLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERE 539
Query: 277 FGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDTTRTL 336
G+ ++ LRSDN Y S F + +SHGI H+ + P TPQ NGVAER +R +V+ R++
Sbjct: 540 TGRKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSM 599
Query: 337 LINAHAPFRFWGDAILIACYLITRMPSSVLGNEIPYSLLFPKDPLYVVPLRVFGSTCFAH 396
L A P FWG+A+ ACYLI R PS L EIP + K+ Y L+VFG FAH
Sbjct: 600 LRMAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSY-SHLKVFGCRAFAH 658
Query: 397 DLSPDRDKLSARAVKCVFLGYSKTQKGYRCYSPSAHRFYVSKDVTFFEDR---------- 446
R KL +++ C+F+GY + GYR + P + S+DV F E
Sbjct: 659 VPKEQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRESEVRTAADMSEK 718
Query: 447 -------PFFASPTTSGSTTSTTSTTD 466
F P+TS + TS STTD
Sbjct: 719 VKNGIIPNFVTIPSTSNNPTSAESTTD 745
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 147 bits (372), Expect = 1e-34
Identities = 97/291 (33%), Positives = 144/291 (49%), Gaps = 17/291 (5%)
Query: 167 HCRLGHPSLDKL-----KVLVPHLSHLKSLD-----CESCQLGKHVRVSFPSRANKRSMS 216
H R GH S KL K + S L +L+ CE C GK R+ F +K +
Sbjct: 419 HERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQARLPFKQLKDKTHIK 478
Query: 217 -PFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSRCTWITLLKDRS*LFGAFQTFCS*IKT 275
P ++HSDV GP + Y+V F+D F+ L+K +S +F FQ F + +
Sbjct: 479 RPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDFVAKSEA 538
Query: 276 QFGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDTTRT 335
F + L DN + Y S F GI + + PHTPQ NGV+ER R + + RT
Sbjct: 539 HFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITEKART 598
Query: 336 LLINAHAPFRFWGDAILIACYLITRMPSSVL--GNEIPYSLLFPKDPLYVVPLRVFGSTC 393
++ A FWG+A+L A YLI R+PS L ++ PY + K P Y+ LRVFG+T
Sbjct: 599 MVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKP-YLKHLRVFGATV 657
Query: 394 FAHDLSPDRDKLSARAVKCVFLGYSKTQKGYRCYSPSAHRFYVSKDVTFFE 444
+ H + + K ++ K +F+GY G++ + +F V++DV E
Sbjct: 658 YVH-IKNKQGKFDDKSFKSIFVGYE--PNGFKLWDAVNEKFIVARDVVVDE 705
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 91.7 bits (226), Expect = 1e-17
Identities = 100/433 (23%), Positives = 173/433 (39%), Gaps = 76/433 (17%)
Query: 66 LNITDANGYKAQVTGIGQASPLPSLSLNY---------VLFISGSPFNLISISKLTQSLN 116
+NI DA + IG +L N+ L ++L+S+S+L N
Sbjct: 480 INIVDAQKQDIPINAIG------NLHFNFQNGTKTSIKALHTPNIAYDLLSLSELANQ-N 532
Query: 117 CSITFSSDSFLIQDRSTGKTIGT*FES*GLYYL--------HSHPSTICGVSASPDI--- 165
+ F+ ++ +RS G + + Y+L H TI V+ S +
Sbjct: 533 ITACFTRNTL---ERSDGTVLAPIVKHGDFYWLSKKYLIPSHISKLTINNVNKSKSVNKY 589
Query: 166 ----IHCRLGHPSLDKL-----KVLVPHL-------SHLKSLDCESCQLGK---HVRVSF 206
IH LGH + + K V +L S+ + C C +GK H V
Sbjct: 590 PYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQCPDCLIGKSTKHRHVKG 649
Query: 207 PSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSRCTWITLLKDR--S*LFG 264
+ S PF +H+D++GP Y+++F D+ +R W+ L DR +
Sbjct: 650 SRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILN 709
Query: 265 AFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAER 324
F + + IK QF + +++ D Y + + + F + GI + + +GVAER
Sbjct: 710 VFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAER 769
Query: 325 KHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLGNEIPYSLLFPKDP---- 380
+R L++ RTLL + P W A+ S+++ N SL+ PK+
Sbjct: 770 LNRTLLNDCRTLLHCSGLPNHLWFSAVEF---------STIIRN----SLVSPKNDKSAR 816
Query: 381 -------LYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLGYSKTQKGYRCYSPSAHR 433
L + + FG ++ +PD K+ R + L S+ GY Y PS +
Sbjct: 817 QHAGLAGLDITTILPFGQPVIVNNHNPD-SKIHPRGIPGYALHPSRNSYGYIIYLPSLKK 875
Query: 434 FYVSKDVTFFEDR 446
+ + +D+
Sbjct: 876 TVDTTNYVILQDK 888
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 85.5 bits (210), Expect = 7e-16
Identities = 87/377 (23%), Positives = 151/377 (39%), Gaps = 46/377 (12%)
Query: 193 CESCQLGK---HVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSR 249
C C +GK H + + S PF +H+D++GP Y+++F D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 250 CTWITLLKDR--S*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGII 307
W+ L DR + F T + IK QF ++ +++ D Y + + + F+ +GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 308 HQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLG 367
+ + +GVAER +R L+D RT L + P W AI S+++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF---------STIVR 380
Query: 368 NEIPYSLLFPKDP-----------LYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLG 416
N SL PK L + L FG +D +P+ K+ R + L
Sbjct: 381 N----SLASPKSKKSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALH 435
Query: 417 YSKTQKGYRCYSPSAHRFYVSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIP 476
S+ GY Y PS + + + + + + T +T S
Sbjct: 436 PSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRLTAS------ 489
Query: 477 LFEPFVSTQNPPQSQGNPEFRQYGITYELR-HVEAPE---TAPIDSNDSTPKTPATNSSD 532
++ F+++ QS + ++ H E P + + DSTP P+T++ D
Sbjct: 490 -YQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTP--PSTHTED 546
Query: 533 SGIVP---VSSPAAVPP 546
S V + +P V P
Sbjct: 547 SKRVSKTNIRAPREVDP 563
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 85.5 bits (210), Expect = 7e-16
Identities = 87/377 (23%), Positives = 151/377 (39%), Gaps = 46/377 (12%)
Query: 193 CESCQLGK---HVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSR 249
C C +GK H + + S PF +H+D++GP Y+++F D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 250 CTWITLLKDR--S*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGII 307
W+ L DR + F T + IK QF ++ +++ D Y + + + F+ +GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 308 HQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLG 367
+ + +GVAER +R L+D RT L + P W AI S+++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF---------STIVR 380
Query: 368 NEIPYSLLFPKDP-----------LYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLG 416
N SL PK L + L FG +D +P+ K+ R + L
Sbjct: 381 N----SLASPKSKKSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALH 435
Query: 417 YSKTQKGYRCYSPSAHRFYVSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIP 476
S+ GY Y PS + + + + + + T +T S
Sbjct: 436 PSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRLTAS------ 489
Query: 477 LFEPFVSTQNPPQSQGNPEFRQYGITYELR-HVEAPE---TAPIDSNDSTPKTPATNSSD 532
++ F+++ QS + ++ H E P + + DSTP P+T++ D
Sbjct: 490 -YQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTP--PSTHTED 546
Query: 533 SGIVP---VSSPAAVPP 546
S V + +P V P
Sbjct: 547 SKRVSKTNIRAPREVDP 563
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 85.5 bits (210), Expect = 7e-16
Identities = 87/377 (23%), Positives = 151/377 (39%), Gaps = 46/377 (12%)
Query: 193 CESCQLGK---HVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSR 249
C C +GK H + + S PF +H+D++GP Y+++F D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 250 CTWITLLKDR--S*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGII 307
W+ L DR + F T + IK QF ++ +++ D Y + + + F+ +GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 308 HQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLG 367
+ + +GVAER +R L+D RT L + P W AI S+++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF---------STIVR 380
Query: 368 NEIPYSLLFPKDP-----------LYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLG 416
N SL PK L + L FG +D +P+ K+ R + L
Sbjct: 381 N----SLASPKSKKSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALH 435
Query: 417 YSKTQKGYRCYSPSAHRFYVSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIP 476
S+ GY Y PS + + + + + + T +T S
Sbjct: 436 PSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRLTAS------ 489
Query: 477 LFEPFVSTQNPPQSQGNPEFRQYGITYELR-HVEAPE---TAPIDSNDSTPKTPATNSSD 532
++ F+++ QS + ++ H E P + + DSTP P+T++ D
Sbjct: 490 -YQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTP--PSTHTED 546
Query: 533 SGIVP---VSSPAAVPP 546
S V + +P V P
Sbjct: 547 SKRVSKTNIRAPREVDP 563
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 85.5 bits (210), Expect = 7e-16
Identities = 87/377 (23%), Positives = 151/377 (39%), Gaps = 46/377 (12%)
Query: 193 CESCQLGK---HVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSR 249
C C +GK H + + S PF +H+D++GP Y+++F D+ ++
Sbjct: 637 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 696
Query: 250 CTWITLLKDR--S*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGII 307
W+ L DR + F T + IK QF ++ +++ D Y + + + F+ +GI
Sbjct: 697 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 756
Query: 308 HQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLG 367
+ + +GVAER +R L+D RT L + P W AI S+++
Sbjct: 757 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF---------STIVR 807
Query: 368 NEIPYSLLFPKDP-----------LYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLG 416
N SL PK L + L FG +D +P+ K+ R + L
Sbjct: 808 N----SLASPKSKKSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALH 862
Query: 417 YSKTQKGYRCYSPSAHRFYVSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIP 476
S+ GY Y PS + + + + + + T +T S
Sbjct: 863 PSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRLTAS------ 916
Query: 477 LFEPFVSTQNPPQSQGNPEFRQYGITYELR-HVEAPE---TAPIDSNDSTPKTPATNSSD 532
++ F+++ QS + ++ H E P + + DSTP P+T++ D
Sbjct: 917 -YQSFIASNEIQQSDDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTP--PSTHTED 973
Query: 533 SGIVP---VSSPAAVPP 546
S V + +P V P
Sbjct: 974 SKRVSKTNIRAPREVDP 990
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 85.1 bits (209), Expect = 9e-16
Identities = 90/376 (23%), Positives = 152/376 (39%), Gaps = 44/376 (11%)
Query: 193 CESCQLGK---HVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSR 249
C C +GK H + + S PF +H+D++GP Y+++F D+ ++
Sbjct: 637 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 696
Query: 250 CTWITLLKDR--S*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGII 307
W+ L DR + F T + IK QF ++ +++ D Y + + + F+ +GI
Sbjct: 697 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 756
Query: 308 HQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLG 367
+ + +GVAER +R L+D RT L + P W AI S+++
Sbjct: 757 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF---------STIVR 807
Query: 368 NEIPYSLLFPKDP-----------LYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLG 416
N SL PK L + L FG +D +P+ K+ R + L
Sbjct: 808 N----SLASPKSKKSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALH 862
Query: 417 YSKTQKGYRCYSPSAHRFYVSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIP 476
S+ GY Y PS + + + + + + T +T S+ I
Sbjct: 863 PSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRLTASYQSFIA 922
Query: 477 LFEPFVSTQNPPQSQGNPEFRQYGITYELRHVEAPE---TAPIDSNDSTPKTPATNSSDS 533
E + N + + +F+ EL H E P + + DSTP P+T++ DS
Sbjct: 923 SNE--IQESNDLNIESDHDFQS---DIEL-HPEQPRNVLSKAVSPTDSTP--PSTHTEDS 974
Query: 534 GIVP---VSSPAAVPP 546
V + +P V P
Sbjct: 975 KRVSKTNIRAPREVDP 990
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 84.3 bits (207), Expect = 2e-15
Identities = 89/375 (23%), Positives = 154/375 (40%), Gaps = 42/375 (11%)
Query: 193 CESCQLGK---HVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSR 249
C C +GK H + + S PF +H+D++GP Y+++F D+ ++
Sbjct: 210 CPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTK 269
Query: 250 CTWITLLKDR--S*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGII 307
W+ L DR + F T + IK QF ++ +++ D Y + + + F+ +GI
Sbjct: 270 FRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGIT 329
Query: 308 HQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLG 367
+ + +GVAER +R L+D RT L + P W AI S+++
Sbjct: 330 PCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF---------STIVR 380
Query: 368 NEIPYSLLFPKDP-----------LYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFLG 416
N SL PK L + L FG +D +P+ K+ R + L
Sbjct: 381 N----SLASPKSKKSARQHAGLAGLDISTLLPFGQPVIVNDHNPN-SKIHPRGIPGYALH 435
Query: 417 YSKTQKGYRCYSPSAHRFYVSKDVTFFEDRPFFASPTTSGSTTSTTSTTDVTTSHVMPIP 476
S+ GY Y PS + + + + + + T +T S+ I
Sbjct: 436 PSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRLTASYHSFIA 495
Query: 477 LFEPFVSTQNPPQSQGNPEFRQYGITY--ELRHVEAPETAPIDSNDSTPKTPATNSSDSG 534
E + N + + +F+ + +LR+V + +P DSTP P+T++ DS
Sbjct: 496 SNE--IQQSNDLNIESDHDFQSDIELHPEQLRNVLSKAVSP---TDSTP--PSTHTEDSK 548
Query: 535 IVP---VSSPAAVPP 546
V + +P V P
Sbjct: 549 RVSKTNIRAPREVDP 563
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 61.6 bits (148), Expect = 1e-08
Identities = 62/292 (21%), Positives = 119/292 (40%), Gaps = 22/292 (7%)
Query: 167 HCRLGHPSLDKLKVLVPHLSHLKSLD---------CESCQLGKHV-RVSFPSRANKRSMS 216
H R+GH + +++ + H + +SLD C++C++ K R + N S
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 217 --PFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSR--CTWITLLKDRS*LFGAFQTFCS* 272
P + D++GP S+ +Y + +D+ +R T K+ + +
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQY 681
Query: 273 IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDT 332
++TQF + +R + SD + + + S GI H + NG AER R ++
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITD 741
Query: 333 TRTLLINAHAPFRFWGDAILIACYLITRMPSSVLGNEIPYSLLFPKDPLYVVPLRVFGST 392
TLL ++ +FW A+ A + + G ++P + + P V +R+
Sbjct: 742 ATTLLRQSNLRVKFWEYAVTSATNIRNYLEHKSTG-KLPLKAI-SRQP---VTVRLMSFL 796
Query: 393 CFAHD---LSPDRDKLSARAVKCVFLGYSKTQKGYRCYSPSAHRFYVSKDVT 441
F + + KL + + L GY+ + PS ++ S + T
Sbjct: 797 PFGEKGIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFFIPSKNKIVTSDNYT 848
>POL_SMSAV (P03359) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 294
Score = 42.4 bits (98), Expect = 0.007
Identities = 64/262 (24%), Positives = 91/262 (34%), Gaps = 36/262 (13%)
Query: 236 GYKYYVTFIDDFSRCTWITLLKDRS*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFST 295
G +Y + FID FS W+ ++ T C T + ++L SDN + +
Sbjct: 26 GNRYLLVFIDTFSG--WVEAFPTKT---ETALTVCKRNSTPL-RIPKVLGSDNGPAFVAQ 79
Query: 296 SFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIAC 355
GI + C + PQ +G ER +R + +T L + + W + +A
Sbjct: 80 VSQGLATQLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLALETGG--KDWVALLPLAL 137
Query: 356 YLITRMPSSVLGNEIPYSLLFPKDPLYVVPLRVFGSTCFAHDLSPDRDKLSARAVKCVFL 415
PS PY +L+ P P+ G T L PD + L L
Sbjct: 138 LRAKNTPSRF--GLTPYEILYGGPP----PILESGGT-----LGPDDNFLPVLFTHLKAL 186
Query: 416 GYSKTQ---KGYRCYSPSA----HRFYVSKDVTFFEDRPFFASPTTSGS-----TTSTTS 463
+TQ + Y P H F V V RP P G TT T
Sbjct: 187 EVVRTQIWDQIKEVYKPGTVAIPHPFQVGDQVLVRRHRPGSLEPRWKGPYLVLLTTPTAV 246
Query: 464 TTD-----VTTSHVMPIPLFEP 480
D V SH+ P P P
Sbjct: 247 KVDGIAAWVHASHLKPAPPSAP 268
>POL_SFV3L (P27401) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1157
Score = 42.0 bits (97), Expect = 0.009
Identities = 60/268 (22%), Positives = 99/268 (36%), Gaps = 29/268 (10%)
Query: 193 CESCQLGKHVRVSFPSRAN-KRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSRCT 251
C+ C + ++ P +R + PF+ D GP S+ GY + + +D +
Sbjct: 860 CKQCLVTNAATLAAPPILRPERPVKPFDKFFIDYIGPLPPSN--GYLHVLVVVDSMTGFV 917
Query: 252 WI--TLLKDRS*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQ 309
W+ T S A S +++ SD + S +F + + GI +
Sbjct: 918 WLYPTKAPSTSATVKALNMLTSIAVP------KVIHSDQGAAFTSATFADWAKNKGIQLE 971
Query: 310 SSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLGNE 369
S P+ PQ +G ERK+ + LL+ A W D +L L S
Sbjct: 972 FSTPYHPQSSGKVERKNSDIKRLLTKLLVGRPAK---WYD-LLPVVQLALNNSYSPSSKY 1027
Query: 370 IPYSLLFPKDPLYVVPLRVFGSTCFAH----DLSPDRDKLSARAVKCVFLGYSKTQKGYR 425
P+ LLF D +T FA+ DLS + + + ++ S R
Sbjct: 1028 TPHQLLFGID----------SNTPFANSDTLDLSREEELSLLQEIRSSLYLPSTPPASIR 1077
Query: 426 CYSPSAHRFYVSKDVTFFEDRPFFASPT 453
+SPS + + RP + PT
Sbjct: 1078 AWSPSVGQLVQERVARPASLRPRWHKPT 1105
>POL_IPMA (P11368) Putative Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 867
Score = 42.0 bits (97), Expect = 0.009
Identities = 21/57 (36%), Positives = 29/57 (50%), Gaps = 4/57 (7%)
Query: 282 RILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDTTRTLLI 338
R+L++DN Y S F F + H + P+ PQ G+ ER HR T +T LI
Sbjct: 706 RLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHR----TLKTYLI 758
>POL_SFV1 (P23074) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1161
Score = 41.6 bits (96), Expect = 0.012
Identities = 60/268 (22%), Positives = 99/268 (36%), Gaps = 29/268 (10%)
Query: 193 CESCQLGKHVRVSFPSRANK-RSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSRCT 251
C+ C + ++ P + + PF+ + D GP S+ GY + + +D +
Sbjct: 858 CKQCLVTNATNLTSPPILRPVKPLKPFDKFYIDYIGPLPPSN--GYLHVLVVVDSMTGFV 915
Query: 252 WI--TLLKDRS*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQ 309
W+ T S A S ++L SD + S++F + GI +
Sbjct: 916 WLYPTKAPSTSATVKALNMLTSIAIP------KVLHSDQGAAFTSSTFADWAKEKGIQLE 969
Query: 310 SSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLGNE 369
S P+ PQ +G ERK+ + LLI A W D +L L S
Sbjct: 970 FSTPYHPQSSGKVERKNSDIKRLLTKLLIGRPAK---WYD-LLPVVQLALNNSYSPSSKY 1025
Query: 370 IPYSLLFPKDPLYVVPLRVFGSTCFAH----DLSPDRDKLSARAVKCVFLGYSKTQKGYR 425
P+ LLF D +T FA+ DLS + + + ++ + R
Sbjct: 1026 TPHQLLFGVD----------SNTPFANSDTLDLSREEELSLLQEIRSSLHQPTSPPASSR 1075
Query: 426 CYSPSAHRFYVSKDVTFFEDRPFFASPT 453
+SPS + + RP + PT
Sbjct: 1076 SWSPSVGQLVQERVARPASLRPRWHKPT 1103
>POL_IPMAI (P12894) Probable Pol polyprotein [Contains: Integrase
(IN); Reverse transcriptase/ribonuclease H (EC 2.7.7.49)
(EC 3.1.26.4) (RT)]
Length = 814
Score = 41.6 bits (96), Expect = 0.012
Identities = 18/48 (37%), Positives = 25/48 (51%)
Query: 282 RILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHL 329
R+L++DN Y S F F + H + P+ PQ G+ ER HR L
Sbjct: 624 RLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 671
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 41.6 bits (96), Expect = 0.012
Identities = 46/188 (24%), Positives = 70/188 (36%), Gaps = 11/188 (5%)
Query: 193 CESCQL-GKHVRVSFPSRANKRSMSPFNIIHSDVWGPSRVSSTLGYKYYVTFIDDFSRCT 251
C+ C + + S P R PF+ D GP S GY Y + +D + T
Sbjct: 650 CQQCLITNASNKASGPILRPDRPQKPFDKFFIDYIGPLPPSQ--GYLYVLVVVDGMTGFT 707
Query: 252 WITLLKDRS*LFGAFQTFCS*IKTQFGKTIRILRSDNAK*YFSTSFNSFMASHGIIHQSS 311
W+ K S T S +++ SD + S++F + GI + S
Sbjct: 708 WLYPTKAPS----TSATVKSLNVLTSIAIPKVIHSDQGAAFTSSTFAEWAKERGIHLEFS 763
Query: 312 CPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLGNEIP 371
P+ PQ ERK+ + LL+ W D + + + S VL P
Sbjct: 764 TPYHPQSGSKVERKNSDIKRLLTKLLVGRPTK---WYDLLPVVQLALNNTYSPVL-KYTP 819
Query: 372 YSLLFPKD 379
+ LLF D
Sbjct: 820 HQLLFGID 827
>IGEB_MOUSE (P03975) IgE-binding protein
Length = 557
Score = 41.6 bits (96), Expect = 0.012
Identities = 18/48 (37%), Positives = 25/48 (51%)
Query: 282 RILRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHL 329
R+L++DN Y S F F + H + P+ PQ G+ ER HR L
Sbjct: 436 RLLKTDNGPAYTSQKFQQFCRQMDVTHLTGLPYNPQGQGIVERAHRTL 483
>POL_JSRV (P31623) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 870
Score = 40.8 bits (94), Expect = 0.020
Identities = 18/46 (39%), Positives = 26/46 (56%)
Query: 284 LRSDNAK*YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHL 329
L++DN Y S SF F S I H++ P+ PQ G+ ER H+ +
Sbjct: 709 LKTDNGPGYTSRSFQRFCLSFQIHHKTGIPYNPQGQGIVERAHQRI 754
>POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1165
Score = 40.8 bits (94), Expect = 0.020
Identities = 74/326 (22%), Positives = 113/326 (33%), Gaps = 37/326 (11%)
Query: 175 LDKLKVLVPHLSHLK---SLDCESCQLGKHVRVSFPSRANKRSMSPFNIIHSDVWGPSRV 231
+++ +L+P+L + C++C + V + +R P ++ +V
Sbjct: 831 VNRTSLLIPNLQSAVREVTSQCQACAMTNAVTTYRETGKRQRGDRPG--VYWEVDFTEIK 888
Query: 232 SSTLGYKYYVTFIDDFSRCTWITLLKDRS*LFGAFQTFCS*IKTQFGKTIRILRSDNAK* 291
G KY + FID FS K + L + I +FG ++L SDN
Sbjct: 889 PGRYGNKYLLVFIDTFSGWVEAFPTKTETALI-VCKKILEEILPRFGIP-KVLGSDNGPA 946
Query: 292 YFSTSFNSFMASHGIIHQSSCPHTPQQNGVAERKHRHLVDTTRTLLINAHAPFRFWGDAI 351
+ + GI + C + PQ +G ER +R + +T L + + W +
Sbjct: 947 FVAQVSQGLATQLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLALETGG--KDWVTLL 1004
Query: 352 LIACYLITRMPSSVLGNEIPYSLLFPKDPLYVVPLRVFGSTCFAHDLSPDRDKLSARAVK 411
+A P PY +L+ P P+ G T L PD L
Sbjct: 1005 PLALLRARNTPGRF--GLTPYEILYGGPP----PILESGET-----LGPDDRFLPVLFTH 1053
Query: 412 CVFLGYSKTQ---KGYRCYSPSA----HRFYVSKDVTFFEDRPFFASPTTSGS-----TT 459
L +TQ + Y P H F V V RP P G TT
Sbjct: 1054 LKALEIVRTQIWDQIKEVYKPGTVTIPHPFQVGDQVLVRRHRPSSLEPRWKGPYLVLLTT 1113
Query: 460 STTSTTD-----VTTSHVMPIPLFEP 480
T D V SH+ P P P
Sbjct: 1114 PTAVKVDGIAAWVHASHLKPAPPSAP 1139
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 40.4 bits (93), Expect = 0.027
Identities = 26/86 (30%), Positives = 40/86 (46%), Gaps = 4/86 (4%)
Query: 326 HRHLVDTTRTLLINAHAPFRFWGDAILIACYLITRMPSSVLGNEIPYSLLFPKDPLYVVP 385
+R +++ R++L P F DA A ++I + PS+ + +P + F P Y
Sbjct: 2 NRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSY- 60
Query: 386 LRVFGSTCFAHDLSPDRDKLSARAVK 411
LR FG + H D KL RA K
Sbjct: 61 LRRFGCVAYIH---CDEGKLKPRAKK 83
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.342 0.151 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 117,130,807
Number of Sequences: 164201
Number of extensions: 4693969
Number of successful extensions: 19017
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 49
Number of HSP's that attempted gapping in prelim test: 18180
Number of HSP's gapped (non-prelim): 322
length of query: 1105
length of database: 59,974,054
effective HSP length: 121
effective length of query: 984
effective length of database: 40,105,733
effective search space: 39464041272
effective search space used: 39464041272
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 71 (32.0 bits)
Medicago: description of AC147007.6


