

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.3 + phase: 0 /pseudo
(651 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
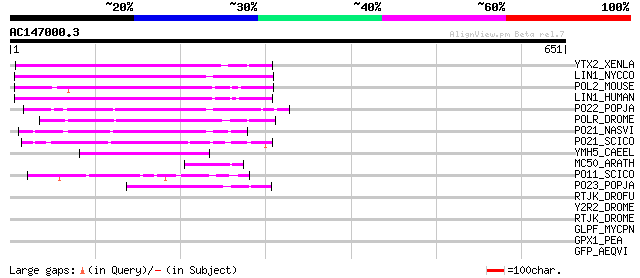
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 129 2e-29
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 124 1e-27
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 115 3e-25
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 103 1e-21
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 81 8e-15
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 72 5e-12
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 69 3e-11
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 61 1e-08
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 57 1e-07
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 52 6e-06
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 50 2e-05
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 49 5e-05
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 39 0.056
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 35 0.62
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 34 1.1
GLPF_MYCPN (P75071) Probable glycerol uptake facilitator protein 33 3.1
GPX1_PEA (O24296) Phospholipid hydroperoxide glutathione peroxid... 32 6.8
GFP_AEQVI (P42212) Green fluorescent protein 31 8.9
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 129 bits (325), Expect = 2e-29
Identities = 82/303 (27%), Positives = 155/303 (51%), Gaps = 9/303 (2%)
Query: 7 LSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFH 66
+S R L P + +E+ QA+ KSPG +G++ F + FWD + DF + L +
Sbjct: 437 VSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAF 496
Query: 67 RNGKLIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSD 126
+ G+L ++L+PK + + + ++ P+SL+ YK++AK ++ RL++V+ V+
Sbjct: 497 KKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHP 556
Query: 127 SQSAFIKGN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNF 186
QS + G I D + + +++ ARR L +D EKA+D VD +YL + +F
Sbjct: 557 DQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSF 616
Query: 187 PTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLM-KQ 245
+ ++ +A+ V +N + RG+RQG PLS L+ LA E F L+ K+
Sbjct: 617 GPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRKR 676
Query: 246 MAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNF 305
+ G LV + ++ + +ADD +++ + +++ + ++ S ++N+
Sbjct: 677 LTG-------LVLKEPDMRVVLSAYADDVILVAQ-DLVDLERAQECQEVYAAASSARINW 728
Query: 306 HKS 308
KS
Sbjct: 729 SKS 731
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 124 bits (310), Expect = 1e-27
Identities = 84/307 (27%), Positives = 147/307 (47%), Gaps = 10/307 (3%)
Query: 6 KLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDF 65
+LS +E L +P S E+ + + KSPGP+G + F + F + + + +
Sbjct: 439 RLSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNI 498
Query: 66 HRNGKLIKGVNSTFIALIPKVN-NPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVV 124
+ G L I LIPK +P R +Y PISL+ K+L K+L NR++ I ++
Sbjct: 499 EKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKII 558
Query: 125 SDSQSAFIKGN*ILDGILIANEVVDDARRM-DKELLLFRVDFEKAYDSVDLKYLDMVMVT 183
Q FI G+ I + V+ ++ +K+ ++ +D EKA+D++ ++ +
Sbjct: 559 HHDQVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMILSIDAEKAFDNIQHPFMIRTLKK 618
Query: 184 MNFPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLM 243
+ + K I A +++NG ++ F + G RQG PLSP L FN++M
Sbjct: 619 IGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLL-------FNIVM 671
Query: 244 KQMAGAQLFNGFLVG-RVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLK 302
+ +A A + G +G + FADD ++ E + + + V+ + VSG K
Sbjct: 672 EVLAIAIREEKAIKGIHIGSEEIKLSLFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYK 731
Query: 303 VNFHKSM 309
+N HKS+
Sbjct: 732 INTHKSV 738
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 115 bits (289), Expect = 3e-25
Identities = 91/310 (29%), Positives = 151/310 (48%), Gaps = 16/310 (5%)
Query: 6 KLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVD- 64
KL+ + +L P S +E++ + + KSPGP+G S F + F K+D + L
Sbjct: 466 KLNQDQVDHLNSPISPKEIEAVINSLPTKKSPGPDGFSAEFYQTF----KEDLIPILHKL 521
Query: 65 FHR---NGKLIKGVNSTFIALIPKVN-NPQRLNDYMPISLVGCLYKVLAKVLANRLRNVI 120
FH+ G L I LIPK +P ++ ++ PISL+ K+L K+LANR++ I
Sbjct: 522 FHKIEVEGTLPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHI 581
Query: 121 GSVVSDSQSAFIKGN*ILDGILIANEVVDDARRM-DKELLLFRVDFEKAYDSVDLKYLDM 179
+++ Q FI G I + V+ ++ DK ++ +D EKA+D + ++
Sbjct: 582 KAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLKDKNHMIISLDAEKAFDKIQHPFMIK 641
Query: 180 VMVTMNFPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGF 239
V+ + I A + VNG +E ++ G RQG PLSP+LF + E
Sbjct: 642 VLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLE-- 699
Query: 240 NVLMKQMAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVS 299
VL + + + G +G+ EV ++ L ADD ++ + R + ++ F EV
Sbjct: 700 -VLARAIRQQKEIKGIQIGK-EEVKISLL--ADDMIVYISDPKNSTRELLNLINSFGEVV 755
Query: 300 GLKVNFHKSM 309
G K+N +KSM
Sbjct: 756 GYKINSNKSM 765
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 103 bits (258), Expect = 1e-21
Identities = 78/305 (25%), Positives = 144/305 (46%), Gaps = 8/305 (2%)
Query: 6 KLSLREAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDF 65
+L+ E +L +P + E++ + + KSPGP G + F +R+ + + ++
Sbjct: 439 RLNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSI 498
Query: 66 HRNGKLIKGVNSTFIALIPKVN-NPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVV 124
+ G L I LIPK + + ++ PISL+ K+L K+LAN+++ I ++
Sbjct: 499 EKEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLI 558
Query: 125 SDSQSAFIKGN*ILDGILIANEVVDDARRM-DKELLLFRVDFEKAYDSVDLKYLDMVMVT 183
Q FI I + ++ R D ++ +D EKA+D + ++ +
Sbjct: 559 HHDQVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMIISIDAEKAFDKIQQPFMLKPLNK 618
Query: 184 MNFPTLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLM 243
+ + K I A +++NG +E ++ G RQG PLSP L + E VL
Sbjct: 619 LGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLLPNIVLE---VLA 675
Query: 244 KQMAGAQLFNGFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKV 303
+ + + G +G+ EV L+ FADD ++ E ++ +++ ++ F +VSG K+
Sbjct: 676 RAIRQEKEIKGIQLGK-EEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKI 732
Query: 304 NFHKS 308
N KS
Sbjct: 733 NVQKS 737
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 81.3 bits (199), Expect = 8e-15
Identities = 72/313 (23%), Positives = 143/313 (45%), Gaps = 20/313 (6%)
Query: 17 KPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVN 76
+P + EE++ A+ +PG +G++ I R + +F+Q + G +
Sbjct: 4 RPIAREEIQCAIKGWKP-SAPGSDGLTVQAITR--TRLPRNFVQLHL---LRGHVPTPWT 57
Query: 77 STFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKGN* 136
+ LIPK + + +++ PI++ L ++L ++LA RL + + +Q + + +
Sbjct: 58 AMRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARIDG 115
Query: 137 ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKWISE 196
L L+ + + R K + +D KA+D+V + + + +I+
Sbjct: 116 TLVNSLLLDTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYITG 175
Query: 197 CIGTAKAAVLVN-GCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGF 255
+ + + V G + I RG++QGDPLSPFL FN ++ ++ +
Sbjct: 176 SLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFL-------FNAVLDELLCSLQSTPG 228
Query: 256 LVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSMLTG*IY 315
+ G +GE + L FADD L++ + L ++ V F + G+ +N KS+ I
Sbjct: 229 IGGTIGEEKIPVLAFADDLLLLEDNDVLLPTTLATVANFF-RLRGMSLNAKKSVS---IS 284
Query: 316 LSLGCGICIELSK 328
++ G+CI +K
Sbjct: 285 VAASGGVCIPRTK 297
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 72.0 bits (175), Expect = 5e-12
Identities = 67/277 (24%), Positives = 114/277 (40%), Gaps = 15/277 (5%)
Query: 36 SPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVNSTFIALIPKVNNPQRLNDY 95
SPGP+GI+ + + M ++ G L + IPK +R D+
Sbjct: 359 SPGPDGITPKSAREVPSGIMLRIMNLILWC---GNLPHSIRLARTVFIPKTVTAKRPQDF 415
Query: 96 MPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKGN*ILDGILIANEVVDDARRMD 155
PIS+ L + L +LA RL + I Q F+ + D I + V+ + +
Sbjct: 416 RPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDGCADNATIVDLVLRHSHKHF 473
Query: 156 KELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKWISECIGTAKAAVLVNGCPMEEF 215
+ + +D KA+DS+ + + P + ++ ++ +G EEF
Sbjct: 474 RSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSEEF 533
Query: 216 SIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLVGRVGEVNLTHLQFADDTL 275
RG++QGDPLSP LF L + + GA +VG FADD +
Sbjct: 534 VPARGVKQGDPLSPILFNLVMDRLLRTLPSEIGA---------KVGNAITNAAAFADDLV 584
Query: 276 IIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHKSMLTG 312
+ E + + ++ + + F + GLK+N K G
Sbjct: 585 LFAE-TRMGLQVLLDKTLDFLSIVGLKLNADKCFTVG 620
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 69.3 bits (168), Expect = 3e-11
Identities = 66/270 (24%), Positives = 120/270 (44%), Gaps = 19/270 (7%)
Query: 11 EAGNLTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGK 70
E NL +P S++E+K+ + + GP+G++ + +K F + +G+
Sbjct: 311 EIKNLWRPISNDEIKEV--EACKRTAAGPDGMTTTAWNSIDECIKSLFNMIMY----HGQ 364
Query: 71 LIKGVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSA 130
+ + LIPK + P+S+ + ++LANR+ ++ Q A
Sbjct: 365 CPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRA 422
Query: 131 FIKGN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLW 190
FI + + + + + ++ +AR K L + +D +KA+DSV+ + + + P
Sbjct: 423 FIVADGVAENTSLLSAMIKEARMKIKGLYIAILDVKKAFDSVEHRSILDALRRKKLPLEM 482
Query: 191 QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQ 250
+ +I +K + V RG+RQGDPLSP L FN +M +
Sbjct: 483 RNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLL-------FNCVMDAVLRRL 535
Query: 251 LFN-GFLVGRVGEVNLTHLQFADDTLIIGE 279
N GFL +G + L FADD +++ E
Sbjct: 536 PENTGFL---MGAEKIGALVFADDLVLLAE 562
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 60.8 bits (146), Expect = 1e-08
Identities = 69/298 (23%), Positives = 130/298 (43%), Gaps = 26/298 (8%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVD-FHRNGKLIK 73
L P S E+K A + K GP+G++ R W+ + D + + L + F G++
Sbjct: 155 LWDPVSLIEIKSA--RASNEKGAGPDGVT----PRSWNALDDRYKRLLYNIFVFYGRVPS 208
Query: 74 GVNSTFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIK 133
+ + PK+ + P+S+ + + K+LA R V + Q+A++
Sbjct: 209 PIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILARRF--VSCYTYDERQTAYLP 266
Query: 134 GN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKW 193
+ + + + ++ +A+R+ KEL + +D KA++SV L + P +
Sbjct: 267 IDGVCINVSMLTAIIAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCPPGVVDY 326
Query: 194 ISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFN 253
I++ + G E SI G+ QGDPLS LF LA + ++ + F+
Sbjct: 327 IADMYNNVITEMQFEG-KCELASILAGVYQGDPLSGPLFTLA---YEKALRALNNEGRFD 382
Query: 254 GFLVGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEV---SGLKVNFHKS 308
+ +V + ++DD L++ + V ++ L F E GL++N KS
Sbjct: 383 ------IADVRVNASAYSDDGLLLA----MTVIGLQHNLDKFGETLAKIGLRINSRKS 430
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 57.4 bits (137), Expect = 1e-07
Identities = 37/153 (24%), Positives = 66/153 (42%), Gaps = 1/153 (0%)
Query: 83 IPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKGN*ILDGIL 142
IPK NP ++Y PISL +++ +++ +R+R+ ++S Q F+ ++
Sbjct: 673 IPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPSSLV 732
Query: 143 IANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKWISECIGTAK 202
+ + + +K L + DF KA+D V L + L W E +
Sbjct: 733 RSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLHLRT 792
Query: 203 AAVLVNG-CPMEEFSIERGLRQGDPLSPFLFLL 234
+V +N + I G+ QG P LF+L
Sbjct: 793 FSVKINKFVSSNAYPISSGVPQGSVSGPLLFIL 825
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 51.6 bits (122), Expect = 6e-06
Identities = 28/69 (40%), Positives = 37/69 (53%), Gaps = 1/69 (1%)
Query: 206 LVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLFNGFLVGRVGEVNL 265
++NG P + RGLRQGDPLSP+LF+L E + L ++ G V +
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSN-NSPRI 71
Query: 266 THLQFADDT 274
HL FADDT
Sbjct: 72 NHLLFADDT 80
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 49.7 bits (117), Expect = 2e-05
Identities = 65/270 (24%), Positives = 110/270 (40%), Gaps = 38/270 (14%)
Query: 22 EEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDD----FMQFLVDFHRNGKLIKGVNS 77
+EV +V C KSPGP+GI ++ W + + + Q L++ + K
Sbjct: 408 DEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIASLV 467
Query: 78 TFIALIPKVNNPQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSV-VSDSQSAFIKGN* 136
+ L+ ++ + Y PI L+ L KVL ++ RL + V VS Q AF G
Sbjct: 468 ILLKLLDRIRSDP--GSYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKS 525
Query: 137 ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVM----VTMNFPTLWQK 192
D V+ + K ++ +DF+ A+D +L +L M++ N LW
Sbjct: 526 TEDAWRCVQRHVECSEM--KYVIGLNIDFQGAFD--NLGWLSMLLKLDEAQSNEFGLWMS 581
Query: 193 WISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLFLLAAEGFNVLMKQMAGAQLF 252
+ + V + + RG QG P ++ L +M ++ A +
Sbjct: 582 YFG-----GRKVYYVGKTGIVRKDVTRGCPQGSKSGPAMWKL-------VMNELLLALVA 629
Query: 253 NGFLVGRVGEVNLTHLQFADD-TLIIGEKS 281
GF + + FADD T++IG S
Sbjct: 630 AGFFI----------VAFADDGTIVIGANS 649
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 48.5 bits (114), Expect = 5e-05
Identities = 42/171 (24%), Positives = 71/171 (40%), Gaps = 10/171 (5%)
Query: 138 LDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPTLWQKWISEC 197
L I++ + R K + +D KA+D+V + M Q +I
Sbjct: 3 LANIIMLEHYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMST 62
Query: 198 IGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLF-LLAAEGFNVLMKQMAGAQLFNGFL 256
I A ++V G + I G++QGDPLSP LF ++ E L + GA +
Sbjct: 63 ITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASM----- 117
Query: 257 VGRVGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNFHK 307
+ L FADD L++ ++ ++V + A + G+ +N K
Sbjct: 118 ---TPACKIASLAFADDLLLLEDRD-IDVPNSLATTCAYFRTRGMTLNPEK 164
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 38.5 bits (88), Expect = 0.056
Identities = 58/225 (25%), Positives = 97/225 (42%), Gaps = 18/225 (8%)
Query: 18 PFSHEEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLV----DFHRNGKLIK 73
P + EEVK+ V K+PG + + I+ L+ D + +LV R G K
Sbjct: 437 PVTLEEVKELVSKLKPKKAPGEDLLDNRTIR----LLPDQALLYLVLIFNSILRVGYFPK 492
Query: 74 GVNSTFIALIPKVNN-PQRLNDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFI 132
+ I +I K P ++ Y P SL+ L K+L +++ NR+ + V+ + F
Sbjct: 493 ARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRI--LTSEEVTRAIPKFQ 550
Query: 133 KGN*ILDGI-----LIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFP 187
G + G + N ++ + + F +D ++A+D V L ++ P
Sbjct: 551 FGFRLQHGTPEQLHRVVNFALEALEKKEYAGSCF-LDIQQAFDRVWHPGLLYKAKSLLSP 609
Query: 188 TLWQKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLF 232
L+Q S G K +V +GC IE G+ QG L P L+
Sbjct: 610 QLFQLIKSFWEG-RKFSVTADGCRSSVKFIEAGVPQGSVLGPTLY 653
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 35.0 bits (79), Expect = 0.62
Identities = 51/217 (23%), Positives = 84/217 (38%), Gaps = 18/217 (8%)
Query: 23 EVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRNGKLIKGVNSTFIAL 82
EV V S +SPG +GI+ K W + + R G +
Sbjct: 438 EVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRVVS 497
Query: 83 IPKVNNPQRL--NDYMPISLVGCLYKVLAKVLANRLRNVIGSVVSDSQSAFIKGN*ILDG 140
+ K + + + Y I L+ KVL ++ NR+R V+ Q F +G + D
Sbjct: 498 LLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPEGCR-WQFGFRQGRCVEDA 556
Query: 141 ILIANEVVDDARRMDKELLLFRVDFEKAYDSVD-----LKYLDMVMVTMNFPTLWQKWIS 195
V + + +L VDF+ A+D+V+ + D+ M LWQ + S
Sbjct: 557 WRHVKSSVGASAA--QYVLGTFVDFKGAFDNVEWSAALSRLADLGCREMG---LWQSFFS 611
Query: 196 ECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLSPFLF 232
+ AV+ + E + RG QG PF++
Sbjct: 612 -----GRRAVIRSSSGTVEVPVTRGCPQGSISGPFIW 643
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 34.3 bits (77), Expect = 1.1
Identities = 29/100 (29%), Positives = 46/100 (46%), Gaps = 9/100 (9%)
Query: 22 EEVKQAVWDCDSFKSPGPNGISFNFIKRFWDLVKDDFMQFLVDFHRN----GKLIKGVNS 77
EEVK + K+PG + + I+ L+ D +QFL + G K S
Sbjct: 441 EEVKNLIAKLPLKKAPGEDLLDNRTIR----LLPDQALQFLALIFNSVLDVGYFPKAWKS 496
Query: 78 TFIALIPKVNN-PQRLNDYMPISLVGCLYKVLAKVLANRL 116
I +I K P ++ Y P SL+ L K++ +++ NRL
Sbjct: 497 ASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRL 536
>GLPF_MYCPN (P75071) Probable glycerol uptake facilitator protein
Length = 264
Score = 32.7 bits (73), Expect = 3.1
Identities = 29/104 (27%), Positives = 45/104 (42%), Gaps = 15/104 (14%)
Query: 129 SAFIKGN*ILDGILIANEVVDDARRMDKELLLFRVDFEKAYDSVDLKYLDMVMVTMNFPT 188
+AF G +L G+LIAN + + A ++ + LF A K L + V + P
Sbjct: 52 AAFAWGIAVLVGVLIANSLFEGAGNINPAVSLF-----YAVSGTIQKALYPLHVNFSIPL 106
Query: 189 LW----QKWISECIGTAKAAVLVNGCPMEEFSIERGLRQGDPLS 228
LW W+++ G A L+N F + + Q DP S
Sbjct: 107 LWVALLLAWVAQFAGAMLAQALLN------FLFWKHIEQTDPQS 144
>GPX1_PEA (O24296) Phospholipid hydroperoxide glutathione
peroxidase, chloroplast precursor (EC 1.11.1.12) (PHGPx)
Length = 236
Score = 31.6 bits (70), Expect = 6.8
Identities = 26/75 (34%), Positives = 41/75 (54%), Gaps = 20/75 (26%)
Query: 15 LTKPFSHEEVKQAVWDCDSFKSPGP-------NGIS----FNFIKR-----FWDLVKDDF 58
+ +P S+EE+KQ + C FK+ P NG + F+K F D+VK +F
Sbjct: 145 MQEPGSNEEIKQ--FACTKFKAEFPIFDKVDVNGPFTAPVYQFLKSSSGGFFGDIVKWNF 202
Query: 59 MQFLVDFHRNGKLIK 73
+FLVD +NGK+++
Sbjct: 203 EKFLVD--KNGKVVE 215
>GFP_AEQVI (P42212) Green fluorescent protein
Length = 238
Score = 31.2 bits (69), Expect = 8.9
Identities = 15/46 (32%), Positives = 24/46 (51%)
Query: 260 VGEVNLTHLQFADDTLIIGEKSWLNVRSMRAVLMLFEEVSGLKVNF 305
V + L + F +D I+G K N S +M ++ +G+KVNF
Sbjct: 120 VNRIELKGIDFKEDGNILGHKLEYNYNSHNVYIMADKQKNGIKVNF 165
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.344 0.156 0.540
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 72,181,927
Number of Sequences: 164201
Number of extensions: 3005740
Number of successful extensions: 11251
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 11208
Number of HSP's gapped (non-prelim): 26
length of query: 651
length of database: 59,974,054
effective HSP length: 117
effective length of query: 534
effective length of database: 40,762,537
effective search space: 21767194758
effective search space used: 21767194758
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 69 (31.2 bits)
Medicago: description of AC147000.3


