

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147000.1 - phase: 0
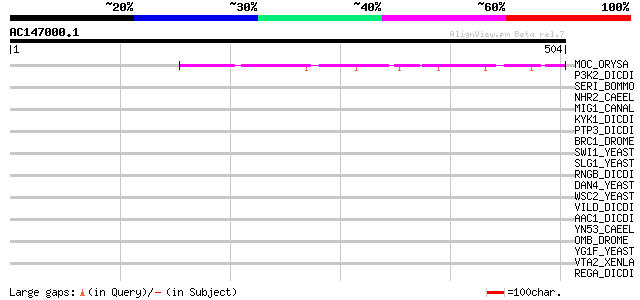
(504 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MOC_ORYSA (Q84MM9) Protein MONOCULM 1 124 4e-28
P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.13... 40 0.014
SERI_BOMMO (P07856) Sericin precursor (Silk gum protein) 39 0.024
NHR2_CAEEL (Q10902) Nuclear hormone receptor family member nhr-2 39 0.031
MIG1_CANAL (Q9Y7G2) Regulatory protein MIG1 38 0.053
KYK1_DICDI (P18160) Non-receptor tyrosine kinase spore lysis A (... 38 0.053
PTP3_DICDI (P54637) Protein-tyrosine phosphatase 3 (EC 3.1.3.48)... 37 0.091
BRC1_DROME (Q01295) Broad-complex core-protein isoforms 1/2/3/4/5 37 0.16
SWI1_YEAST (P09547) Transcription regulatory protein SWI1 (SWI/S... 36 0.27
SLG1_YEAST (P54867) SLG1 protein precursor (Cell wall integrity ... 36 0.27
RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB) 36 0.27
DAN4_YEAST (P47179) Cell wall protein DAN4 precursor 36 0.27
WSC2_YEAST (P53832) Cell wall integrity and stress response comp... 35 0.35
VILD_DICDI (Q8WQ85) Villidin 35 0.45
AAC1_DICDI (P14195) AAC-rich mRNA clone AAC1 protein (Fragment) 35 0.45
YN53_CAEEL (P34587) Hypothetical protein ZC21.3 in chromosome III 35 0.59
OMB_DROME (Q24432) Optomotor-blind protein (Lethal(1)optomotor-b... 35 0.59
YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A... 34 0.77
VTA2_XENLA (P18709) Vitellogenin A2 precursor (VTG A2) [Contains... 34 0.77
REGA_DICDI (Q23917) 3',5'-cyclic-nucleotide phosphodiesterase re... 34 0.77
>MOC_ORYSA (Q84MM9) Protein MONOCULM 1
Length = 441
Score = 124 bits (312), Expect = 4e-28
Identities = 106/385 (27%), Positives = 176/385 (45%), Gaps = 57/385 (14%)
Query: 155 MLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEKTCSFDSTRKMLLKFQ 214
+L ++P GD +L+ +F +AL R++ + + + + S + L F
Sbjct: 76 VLAAAASPRGDAADRLAYHFARALALRVDAKAGHGHVVVGGGAARPASSGA----YLAFN 131
Query: 215 EVSPWTTFGHVAANGAILEALEGNPKLHIIDISNTYCTQWPTLLEALATRSDDT---PHL 271
+++P+ F H+ AN AILEA++G ++HI+D+ + QWP LL+A+A R+D P +
Sbjct: 132 QIAPFLRFAHLTANQAILEAVDGARRVHILDLDAVHGVQWPPLLQAIAERADPALGPPEV 191
Query: 272 RLTTVVTAISGGSVQKVMKEIGSRMEKFARLMGVPFKFKIIF------------------ 313
R+T G+ + + G+R+ FAR + +PF F +
Sbjct: 192 RVT------GAGADRDTLLRTGNRLRAFARSIHLPFHFTPLLLSCATTAPHHVAGTSTGA 245
Query: 314 --SDLRELNLCDLDIKEDEALAINCVNSLHSISGAGNHRDL--FISLLRGLEPRVLTIVE 369
+ L+ DE LA+NCV LH+++G H +L F+ ++ + P V+TI E
Sbjct: 246 AAAASTAAAATGLEFHPDETLAVNCVMFLHNLAG---HDELAAFLKWVKAMSPAVVTIAE 302
Query: 370 EEADLEVCFGSDFVEGFKE----CLRWFRVYFEALDESFSRTSSERLMLEREA-GRGIVD 424
EA G D ++ + + FEAL+ + S ERL +E+E GR I
Sbjct: 303 REAG-GGGGGGDHIDDLPRRVGVAMDHYSAVFEALEATVPPGSRERLAVEQEVLGREIEA 361
Query: 425 LVACDP---YESVERRETAARWRRRLHGGGFNTVSFSDEVCDDVRALLRRY--KEGWSMT 479
V + +ER AAR GF S R LLR + EG+ +
Sbjct: 362 AVGPSGGRWWRGIERWGGAAR------AAGFAARPLSAFAVSQARLLLRLHYPSEGYLVQ 415
Query: 480 SSDGDTGIFLSWKDKPVVWASVWRP 504
+ G FL W+ +P++ S W+P
Sbjct: 416 EARG--ACFLGWQTRPLLSVSAWQP 438
>P3K2_DICDI (P54674) Phosphatidylinositol 3-kinase 2 (EC 2.7.1.137)
(PI3-kinase) (PtdIns-3-kinase) (PI3K)
Length = 1858
Score = 40.0 bits (92), Expect = 0.014
Identities = 29/90 (32%), Positives = 40/90 (44%), Gaps = 7/90 (7%)
Query: 24 NSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSS---SSKQH 80
N+T+T TT++S S + ++E FNN ++NNNN SS +SK
Sbjct: 226 NTTSTTTTTTSILISSSPPPSSSSSSSSNDEQFNN----NNNNNNSNSGGSSRMITSKSQ 281
Query: 81 YYTYPYASTTTITTPNTTYNTINTPTTTDN 110
S T TT TT + PTT N
Sbjct: 282 IKPLIVTSNTAATTTTTTTTNTSAPTTPTN 311
Score = 38.1 bits (87), Expect = 0.053
Identities = 23/101 (22%), Positives = 51/101 (49%)
Query: 21 PSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQH 80
P N+ T+ ++SSS+ ++ + N + NN ++NNNN+ + +++++ +
Sbjct: 159 PICNNLTSSSSSSSTTATTPSPTTTSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN 218
Query: 81 YYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFN 121
+TT+ TT T+ ++P + + S S + + FN
Sbjct: 219 NNNNNNNNTTSTTTTTTSILISSSPPPSSSSSSSSNDEQFN 259
Score = 34.3 bits (77), Expect = 0.77
Identities = 18/37 (48%), Positives = 24/37 (64%)
Query: 6 RLVNFQQQQQQYQPDPSLNSTTTLTTSSSSRSSRQTT 42
+L+ QQQQQ + + S N+ TT T+SSSS SS T
Sbjct: 420 QLIPSQQQQQPTESNSSSNTNTTTTSSSSSSSSSSLT 456
>SERI_BOMMO (P07856) Sericin precursor (Silk gum protein)
Length = 389
Score = 39.3 bits (90), Expect = 0.024
Identities = 31/131 (23%), Positives = 61/131 (45%), Gaps = 5/131 (3%)
Query: 20 DPSLNSTTTLTTSSSSRSSRQTTYHYY-NQQEEDEECFNNFYYMDHNNNNDEDLSSSSSK 78
D S N+ + ++ SS S + TY Y N ++ + D N++N +S S
Sbjct: 152 DASSNTDSNSNSAGSSTSGGRRTYGYSSNSRDGSVSSTGSSSNTDSNSSNAGSSTSGGSS 211
Query: 79 QHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETA 138
TY Y+S + + +TT ++ NT + +++ S + E S S +N+ +
Sbjct: 212 ----TYGYSSNSRDGSVSTTGSSSNTDSNSNSVGSRRSGGSSSHEDSSKSRDENVSTTGS 267
Query: 139 RAFSDNNTNRI 149
+ +D+N+N +
Sbjct: 268 SSNTDSNSNSV 278
Score = 34.7 bits (78), Expect = 0.59
Identities = 27/126 (21%), Positives = 63/126 (49%), Gaps = 2/126 (1%)
Query: 22 SLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHY 81
S ++T + + S SR S ++ H + + DE + ++N++ SS+S +
Sbjct: 229 SSSNTDSNSNSVGSRRSGGSSSHEDSSKSRDENVSTTGSSSNTDSNSNSVGSSTSGGRR- 287
Query: 82 YTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETARAF 141
TY Y+S + + ++T ++ NT + +++ S S + +S +S ++ + +
Sbjct: 288 -TYGYSSNSRDGSVSSTGSSSNTDSNSNSVGSSTSGGSSTYGYSSNSRDGSVSSTGSSSN 346
Query: 142 SDNNTN 147
+D+N+N
Sbjct: 347 TDSNSN 352
>NHR2_CAEEL (Q10902) Nuclear hormone receptor family member nhr-2
Length = 359
Score = 38.9 bits (89), Expect = 0.031
Identities = 22/82 (26%), Positives = 40/82 (47%), Gaps = 2/82 (2%)
Query: 65 NNNNDEDLSSSSSKQHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEF 124
N N +D +SS +++ T+P +S +T T Y+ NTP T + D+ N+
Sbjct: 34 NGNEADDKENSSVRRN--TFPASSASTKKPRGTPYDRTNTPMTVVPQMPQFAPDFSNYNL 91
Query: 125 SGHSWSQNILLETARAFSDNNT 146
+ ++WS+ + DN+T
Sbjct: 92 TMNAWSEFFKKDRCMVCGDNST 113
>MIG1_CANAL (Q9Y7G2) Regulatory protein MIG1
Length = 574
Score = 38.1 bits (87), Expect = 0.053
Identities = 39/147 (26%), Positives = 53/147 (35%), Gaps = 33/147 (22%)
Query: 11 QQQQQQYQPDPSLNSTTTLTTSSSSRSSR-QTTYHYYNQQEEDEEC-------------- 55
QQQQQQ Q P S+T+L + + SR +T H E+
Sbjct: 268 QQQQQQQQQIPKSESSTSLYSDGNKLFSRPNSTLHSLGSSPENNNASGPLSGVPTPSFSN 327
Query: 56 FNNFYYMDHNNNNDEDLSSSSSKQHYYT-----------------YPYASTTTITTPNTT 98
N+++ N+NN ++SSS + P STT T TT
Sbjct: 328 LNDYFQQKSNSNNSRLFNASSSSLSSLSGKIRSSSSTNLAGLQRLTPLTSTTNNTNNTTT 387
Query: 99 YNTINTPTTTDN-YSFSPSHDYFNFEF 124
NT N T + PS N EF
Sbjct: 388 SNTNNNNMTKPSIIPKQPSSTSLNLEF 414
>KYK1_DICDI (P18160) Non-receptor tyrosine kinase spore lysis A (EC
2.7.1.112) (Tyrosine-protein kinase 1)
Length = 1584
Score = 38.1 bits (87), Expect = 0.053
Identities = 34/146 (23%), Positives = 64/146 (43%), Gaps = 26/146 (17%)
Query: 23 LNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHYY 82
+ +TTT +TS SS ++ + N + NN ++NNNN+ + ++S+S
Sbjct: 427 ITTTTTTSTSPSSINNNEDISSNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNSNSS---- 482
Query: 83 TYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETARAFS 142
NT N IN T +N + + +++ N + +S + NI +
Sbjct: 483 -------------NTNNNNINNTTNNNNSNSNNNNNNNNSNSNSNSNNNNI-------NN 522
Query: 143 DNNTNRIQQLMWMLNELSTPYGDTDQ 168
+NN N +++ + S G TD+
Sbjct: 523 NNNNNNNNNNIYLTKKPS--IGSTDE 546
>PTP3_DICDI (P54637) Protein-tyrosine phosphatase 3 (EC 3.1.3.48)
(Protein-tyrosine-phosphate phosphohydrolase 3)
Length = 989
Score = 37.4 bits (85), Expect = 0.091
Identities = 29/132 (21%), Positives = 51/132 (37%), Gaps = 26/132 (19%)
Query: 13 QQQQYQPDPSLNSTTTLTTSSSS----------------------RSSRQTTYHYYNQQE 50
QQQQ Q LN L SS+S +++Q QQ+
Sbjct: 834 QQQQQQSSQQLNDNPPLNMSSNSIKFPPVTSLSSCHLFEDSKNNDNNNKQQQQQQQQQQK 893
Query: 51 EDEECFNNFYYMDHNNNNDEDLSSSSSKQHYYTY----PYASTTTITTPNTTYNTINTPT 106
+++C +++++NNNND + SS + + +S+T N N N
Sbjct: 894 NNQQCSGFSHFLNNNNNNDNNGSSGGGFNGSFLFNSNNSGSSSTNSECSNNNKNNNNNSN 953
Query: 107 TTDNYSFSPSHD 118
+N + + + D
Sbjct: 954 NNNNNNNNKNSD 965
Score = 35.8 bits (81), Expect = 0.27
Identities = 39/145 (26%), Positives = 57/145 (38%), Gaps = 46/145 (31%)
Query: 15 QQYQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDED--- 71
QQ SL+STTT TT++SS + FNN Y +H+NNN+
Sbjct: 247 QQSSSSSSLSSTTTTTTTTSSSLLMP------------QSLFNNSTYNNHHNNNNSSNAG 294
Query: 72 ----LSSSSS--------------------KQHYYTYP-YASTTTITTPNTTYNTINTPT 106
L+ S+S +QH Y +S +T+ N N +NT T
Sbjct: 295 IVGGLNGSTSSLPTQAQVQLQQMQQQMQQHQQHQYKKANLSSLSTVVDNNLNNNPMNTST 354
Query: 107 TT------DNYSFSPSHDYFNFEFS 125
++ +SFS S + N S
Sbjct: 355 SSPAQPNASPFSFSSSSLFSNSSLS 379
Score = 34.7 bits (78), Expect = 0.59
Identities = 32/133 (24%), Positives = 57/133 (42%), Gaps = 3/133 (2%)
Query: 17 YQPDPSLNS--TTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSS 74
Y +P LN +T+ T +S + T +Q E N + NNNN+ S
Sbjct: 16 YTLNPHLNIPISTSTTIPPTSFYANNTPEMIQSQSENTNTNNINNSSSNINNNNNNTPDS 75
Query: 75 SSSKQHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNIL 134
S + P S + N+ N IN TTT+N + + +++ F+ + S ++
Sbjct: 76 MSMSTSLSSSPSVSFNHLDL-NSINNKINNNTTTNNNNNNNNNNDDKFDTNALKLSNTMI 134
Query: 135 LETARAFSDNNTN 147
++ ++NN N
Sbjct: 135 IKNNNNNNNNNNN 147
Score = 31.2 bits (69), Expect = 6.5
Identities = 21/87 (24%), Positives = 39/87 (44%), Gaps = 19/87 (21%)
Query: 8 VNFQQQQQQ-------------------YQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQ 48
+N QQQQQQ + P SL+S S ++ ++ + Q
Sbjct: 831 LNLQQQQQQSSQQLNDNPPLNMSSNSIKFPPVTSLSSCHLFEDSKNNDNNNKQQQQQQQQ 890
Query: 49 QEEDEECFNNFYYMDHNNNNDEDLSSS 75
Q+++ + + F + +NNNN+++ SS
Sbjct: 891 QQKNNQQCSGFSHFLNNNNNNDNNGSS 917
>BRC1_DROME (Q01295) Broad-complex core-protein isoforms 1/2/3/4/5
Length = 727
Score = 36.6 bits (83), Expect = 0.16
Identities = 27/85 (31%), Positives = 40/85 (46%), Gaps = 22/85 (25%)
Query: 24 NSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHYYT 83
N+TTT TT ++S ++ NN ++NNNN+ D S + +
Sbjct: 437 NATTTTTTINNSITN------------------NN----NNNNNNNYDYSLPTKNSNSQK 474
Query: 84 YPYASTTTITTPNTTYNTINTPTTT 108
P +TTT+TTP TT T T T+
Sbjct: 475 TPSPTTTTLTTPTTTTPTRPTAITS 499
>SWI1_YEAST (P09547) Transcription regulatory protein SWI1 (SWI/SNF
complex component SWI1) (Transcription regulatory
protein ADR6) (Regulatory protein GAM3)
Length = 1314
Score = 35.8 bits (81), Expect = 0.27
Identities = 43/192 (22%), Positives = 78/192 (40%), Gaps = 23/192 (11%)
Query: 20 DPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFY-------YMDHNNNNDEDL 72
+P+ N+ +T SS ++ T + N + F NF+ +D NNNN
Sbjct: 37 NPANNTNNNNSTGHSSNTNNNTNNNNTNTGASGVDDFQNFFDPKPFDQNLDSNNNN---- 92
Query: 73 SSSSSKQHYYTYPYASTTTITTPNTTYNTINTPTTT----------DNYSFSPSHDYFNF 122
S+S++ + + AS+T T+P N T + F+ +D N
Sbjct: 93 SNSNNNDNNNSNTVASSTNFTSPTAVVNNAAPANVTGGKAANFIQNQSPQFNSPYDSNNS 152
Query: 123 EFSGHSWS-QNILLETARAFSDNNTNRIQQLMWMLNELSTPYG-DTDQKLSSYFLQALFS 180
+ +S S Q IL + + S N + QQ ++ N + G D ++ +F+ + S
Sbjct: 153 NTNLNSLSPQAILAKNSIIDSSNLPLQAQQQLYGGNNNNNSTGIANDNVITPHFITNVQS 212
Query: 181 RMNDAGDRTYKT 192
++ T T
Sbjct: 213 ISQNSSSSTPNT 224
Score = 32.7 bits (73), Expect = 2.2
Identities = 25/90 (27%), Positives = 41/90 (44%), Gaps = 27/90 (30%)
Query: 58 NFYYMDHNNNNDEDLSSSSSKQHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSH 117
+F+ +++NNNN+ +TTT TT NT N T +N +P++
Sbjct: 2 DFFNLNNNNNNN------------------NTTTTTTTTNNNNTNNNNTNNNN---NPAN 40
Query: 118 DYFNFEFSGHSWSQNILLETARAFSDNNTN 147
+ N +GHS + N ++NNTN
Sbjct: 41 NTNNNNSTGHSSNTN------NNTNNNNTN 64
>SLG1_YEAST (P54867) SLG1 protein precursor (Cell wall integrity and
stress response component 1)
Length = 378
Score = 35.8 bits (81), Expect = 0.27
Identities = 29/90 (32%), Positives = 46/90 (50%), Gaps = 3/90 (3%)
Query: 28 TLTTSSSSRSSRQTTYHY-YNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHYYTYPY 86
T + S S SS TY + Y+ + E + Y +D + N++ SS SS + T
Sbjct: 74 TNPSGSESTSSSCNTYCFGYSSEMCGGEDAYSVYQLDSDTNSNSISSSDSSTES--TSAS 131
Query: 87 ASTTTITTPNTTYNTINTPTTTDNYSFSPS 116
+STT+ TT +TT T +T ++T + S S
Sbjct: 132 SSTTSSTTSSTTSTTSSTTSSTTSSMASSS 161
>RNGB_DICDI (Q7M3S9) Ring finger protein B (Protein rngB)
Length = 943
Score = 35.8 bits (81), Expect = 0.27
Identities = 39/171 (22%), Positives = 67/171 (38%), Gaps = 19/171 (11%)
Query: 8 VNFQQQQQQYQPDPSLNSTT----TLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMD 63
+ F Q Q Q + + N+ T TT++++ ++ + N + E +
Sbjct: 403 LQFNQNQNQNNNNNNNNNNNNNIQTTTTTTTNNNNNNNNNNNNNNNNNNVESNQQQQQIQ 462
Query: 64 HNNNNDEDLSSSSSKQHYYTYPYASTTTITTPNTTYNTINTPTT--------TDNYSFS- 114
H + LS S+S + +S++ ++TP+T T T TT T N S
Sbjct: 463 HQTSPMSVLSRSNSNISLNSLNSSSSSILSTPSTLSTTTTTTTTSHASHTSHTSNRSNGS 522
Query: 115 ----PSHDYFNFEFSGHSWSQNILLETARAFSDNNTNRIQQLMWMLNELST 161
PS FN S H+ + N TN ++L +L EL +
Sbjct: 523 RGGIPSIPPFNGRSSNHNNNNNSNSNNYNNHQQTKTNSAEEL--ILEELKS 571
>DAN4_YEAST (P47179) Cell wall protein DAN4 precursor
Length = 1161
Score = 35.8 bits (81), Expect = 0.27
Identities = 46/190 (24%), Positives = 72/190 (37%), Gaps = 13/190 (6%)
Query: 19 PDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSK 78
P S STT T+++S+ + TT + + SS+S+
Sbjct: 198 PTTSTTSTTPTTSTTSTTPTTSTTSTTPTTSTTPTTSTTSTTSQTSTKSTTPTTSSTSTT 257
Query: 79 QHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETA 138
T P STT+ T P T+ T +T +TT S +P+ + FS S S + ++ T
Sbjct: 258 PTTSTTPTTSTTS-TAPTTS--TTSTTSTTSTISTAPTTSTTSSTFSTSSASASSVISTT 314
Query: 139 RAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALF--SRMNDAGDRTYKTLTTA 196
S + L +T TD SS F S TY + ++
Sbjct: 315 ATTSTTFAS--------LTTPATSTASTDHTTSSVSTTNAFTTSATTTTTSDTYISSSSP 366
Query: 197 SEKTCSFDST 206
S+ T S + T
Sbjct: 367 SQVTSSAEPT 376
>WSC2_YEAST (P53832) Cell wall integrity and stress response
component 2 precursor
Length = 503
Score = 35.4 bits (80), Expect = 0.35
Identities = 39/190 (20%), Positives = 74/190 (38%), Gaps = 10/190 (5%)
Query: 22 SLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHY 81
S +T+T TTSSSS S T + + H++++ S+++S
Sbjct: 127 SSTATSTSTTSSSSTSVSSKTSTKLDTKTSTSSS------ATHSSSSSSTTSTTTSSSET 180
Query: 82 YTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSWSQNILLETARAF 141
T S+++ ++ ++T T T TT+ S S S + S S S+ + +
Sbjct: 181 TT----SSSSSSSSSSTSTTSTTSTTSSTTSTSSSPSTTSSSTSASSSSETSSTQATSSS 236
Query: 142 SDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFLQALFSRMNDAGDRTYKTLTTASEKTC 201
+ + ++ ST G + + Q++ S+ N + T T+
Sbjct: 237 TTSTSSSTSTATVTSTPSSTSIGTSTHYTTRVVTQSVVSQANQQASTIFTTRTSVYATVS 296
Query: 202 SFDSTRKMLL 211
S S+ LL
Sbjct: 297 STSSSTSSLL 306
>VILD_DICDI (Q8WQ85) Villidin
Length = 1704
Score = 35.0 bits (79), Expect = 0.45
Identities = 42/148 (28%), Positives = 55/148 (36%), Gaps = 14/148 (9%)
Query: 11 QQQQQQYQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDE 70
+QQQQQ Q S NSTT S + QT +H N Y ++
Sbjct: 612 EQQQQQQQGSESPNSTT-----PKSSTPTQTRHH--------SVLSRNPSYTSLKSSGGF 658
Query: 71 DLSSSSSKQHYYTYPYASTTTIT-TPNTTYNTINTPTTTDNYSFSPSHDYFNFEFSGHSW 129
S SS +AS +I T +TT T T TTT + S P N S
Sbjct: 659 SKPSPSSSTSTPPSTFASPLSIAQTSSTTTTTTTTTTTTSSSSTPPPLRANNSTSSTPLN 718
Query: 130 SQNILLETARAFSDNNTNRIQQLMWMLN 157
S + ++ N T I L+W N
Sbjct: 719 SSTGIKQSDIVIEGNLTELIPGLLWNSN 746
>AAC1_DICDI (P14195) AAC-rich mRNA clone AAC1 protein (Fragment)
Length = 183
Score = 35.0 bits (79), Expect = 0.45
Identities = 27/96 (28%), Positives = 46/96 (47%), Gaps = 9/96 (9%)
Query: 12 QQQQQYQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDED 71
+QQ+ + + S STTT TT ++ ++ TT N + + N+ N ++
Sbjct: 12 KQQKLLKSNESTTSTTTTTTPITTTTTTTTTTTTPNLSK-----YYNYNLYIEKQNEKQN 66
Query: 72 LSSSSSKQHYYTYPYASTTTITT----PNTTYNTIN 103
L ++ ++ T +TTT TT NTT +TIN
Sbjct: 67 LPTTETETTTITPTLTTTTTTTTTKQIQNTTTSTIN 102
Score = 33.9 bits (76), Expect = 1.0
Identities = 23/59 (38%), Positives = 29/59 (48%), Gaps = 8/59 (13%)
Query: 67 NNDEDLSSSSSKQHYY---TYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNF 122
+ND + S+ +KQ STTT TTP TT T T TTT N S Y+N+
Sbjct: 1 DNDTNSSTRPNKQQKLLKSNESTTSTTTTTTPITTTTTTTTTTTTPNLS-----KYYNY 54
>YN53_CAEEL (P34587) Hypothetical protein ZC21.3 in chromosome III
Length = 321
Score = 34.7 bits (78), Expect = 0.59
Identities = 29/113 (25%), Positives = 49/113 (42%), Gaps = 7/113 (6%)
Query: 9 NFQQQQQQYQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEEC----FNNFYYMDH 64
++Q Q +P TTT +SS + +Q YN Q +++ +NN D
Sbjct: 204 SYQNQMMYNNNNPQNTQTTTFLYASSIQPQQQQQQQQYNNQYNNQQMNSNGYNNQPMYDQ 263
Query: 65 NNNNDEDLSSSSSKQHYY-TYPYASTTTITTPNTTYNTINTPTTTDNYSFSPS 116
+ + + SS+ S Q+ + S + T T NTP ++N +SPS
Sbjct: 264 SYSTQQYGSSTPSPQYSADSISCCSQVSSTCCYQTQQGYNTP--SNNVQYSPS 314
Score = 33.9 bits (76), Expect = 1.0
Identities = 39/145 (26%), Positives = 62/145 (41%), Gaps = 22/145 (15%)
Query: 33 SSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHYYTYPYAST--T 90
SSS + + Y +QQ++ ++ +N Y N + + +Q Y T ++S
Sbjct: 4 SSSSTGQYNQQQYNSQQQQPQQQYNTQQY-----NPQQQQYNPQQQQQYGTTSFSSNGQA 58
Query: 91 TITTPNTTYNTINTPTTTD-NYS-FSPSHDYFNFEFSGHSWSQNILLETARAFSDNNTNR 148
T T Y T+ P+++ YS +PS Y G+S SQN +A S +N R
Sbjct: 59 MYQTGTTNYPTLYQPSSSSPQYSNGAPSTTYSPTSTYGYS-SQNYNPSSA---SSSNVYR 114
Query: 149 IQQLMWMLNELSTPYGDTDQKLSSY 173
ST YG+T Q +Y
Sbjct: 115 ---------NPSTSYGNTQQSNQNY 130
>OMB_DROME (Q24432) Optomotor-blind protein
(Lethal(1)optomotor-blind) (L(1)omb) (Bifid protein)
Length = 972
Score = 34.7 bits (78), Expect = 0.59
Identities = 26/103 (25%), Positives = 42/103 (40%), Gaps = 8/103 (7%)
Query: 16 QYQPDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSS 75
Q P + +S+ T TTS+++ SS + + NN +NNNN+ + +S
Sbjct: 92 QLSPQSNHSSSNTTTTSNTNNSS--------SNNNNNNSTHNNNNNHTNNNNNNNNNTSQ 143
Query: 76 SSKQHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHD 118
H T + T P T P +N S S S++
Sbjct: 144 KQGHHLSTTEEPPSPAGTPPPTIVGLPPIPPPNNNSSSSSSNN 186
>YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A
intergenic region
Length = 551
Score = 34.3 bits (77), Expect = 0.77
Identities = 27/107 (25%), Positives = 54/107 (50%), Gaps = 16/107 (14%)
Query: 24 NSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSKQHYYT 83
+S++ LT+SS S SS +T Y+ ++ +++ SSSSS Y+T
Sbjct: 215 SSSSELTSSSYSSSSSSSTLFSYSSSFSSSSSSSS-------SSSSSSSSSSSSSSSYFT 267
Query: 84 YPYASTTTI----TTPNTTYNTINTPT-----TTDNYSFSPSHDYFN 121
+S+++I + P+ + ++ + PT T+ + S +P+ +Y N
Sbjct: 268 LSTSSSSSIYSSSSYPSFSSSSSSNPTSSITSTSASSSITPASEYSN 314
>VTA2_XENLA (P18709) Vitellogenin A2 precursor (VTG A2) [Contains:
Lipovitellin I; Lipovitellin II; Phosvitin]
Length = 1807
Score = 34.3 bits (77), Expect = 0.77
Identities = 37/166 (22%), Positives = 73/166 (43%), Gaps = 18/166 (10%)
Query: 22 SLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEEC-FNNFYYMDHNNNNDEDLSSSSS--- 77
S +S+++ ++SSSS SS + + N+Q ++++ + + N E SSSSS
Sbjct: 1215 SSSSSSSSSSSSSSSSSSEENRPHKNRQHDNKQAKMQSNQHQQKKNKFSESSSSSSSSSS 1274
Query: 78 -------KQHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDY-FNFEFSGHSW 129
K H Y T T T + +++++ S S Y +F G
Sbjct: 1275 SEMWNKKKHHRNFYDLNFRRTARTKGTEHRGSRLSSSSESSSSSSESAYRHKAKFLGDK- 1333
Query: 130 SQNILLETARAFSDNNTNRIQQLMWMLNELSTPYGDTDQKLSSYFL 175
+L+ T +A ++NT + Q++ + Y + Q++ +Y +
Sbjct: 1334 EPPVLVVTFKAVRNDNTKQGYQMV-----VYQEYHSSKQQIQAYVM 1374
>REGA_DICDI (Q23917) 3',5'-cyclic-nucleotide phosphodiesterase regA
(EC 3.1.4.17) (PDEase regA)
Length = 793
Score = 34.3 bits (77), Expect = 0.77
Identities = 26/112 (23%), Positives = 53/112 (47%), Gaps = 2/112 (1%)
Query: 19 PDPSLNSTTTLTTSSSSRSSRQTTYHYYNQQEEDEECFNNFYYMDHNNNNDEDLSSSSSK 78
P PS +S+ + S+S +S + + ++ NN ++N+NN+E S
Sbjct: 19 PSPSSSSSPSNNDSTSLKSMISGIENLNVHSKGNDNKNNNNNNNNNNSNNNEKQKDIVSL 78
Query: 79 QHYYTYPYASTTTITTPNTTYNTINTPTTTDNYSFSPSHDYFNFE--FSGHS 128
++ + +TTT TT + +N+ N +N + + + N+E +GH+
Sbjct: 79 ENNSSSNNTTTTTTTTTTSNHNSNNNSNNNNNNINNNNINNNNYEPLVNGHN 130
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.131 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 60,727,109
Number of Sequences: 164201
Number of extensions: 2557458
Number of successful extensions: 10486
Number of sequences better than 10.0: 81
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 9973
Number of HSP's gapped (non-prelim): 301
length of query: 504
length of database: 59,974,054
effective HSP length: 115
effective length of query: 389
effective length of database: 41,090,939
effective search space: 15984375271
effective search space used: 15984375271
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC147000.1


