

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146972.3 + phase: 0
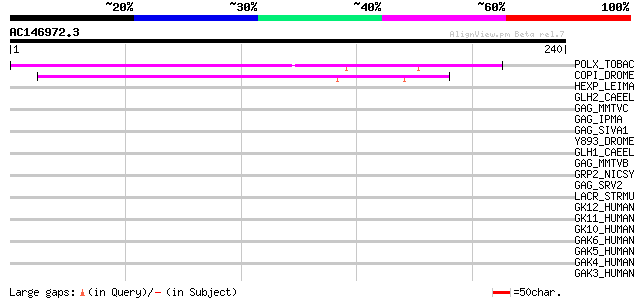
(240 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 139 5e-33
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 66 8e-11
HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding p... 37 0.032
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 37 0.042
GAG_MMTVC (P11284) Gag polyprotein [Contains: Protein p10; Phosp... 37 0.042
GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains: ... 37 0.042
GAG_SIVA1 (P27972) Gag polyprotein [Contains: Core protein p17; ... 36 0.094
Y893_DROME (Q9VV43) Hypothetical protein CG4893 35 0.12
GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-... 35 0.12
GAG_MMTVB (P10258) Gag polyprotein [Contains: Protein p10; Phosp... 35 0.12
GRP2_NICSY (P27484) Glycine-rich protein 2 35 0.16
GAG_SRV2 (P51516) Gag polyprotein (Core polyprotein) [Contains: ... 35 0.21
LACR_STRMU (P26422) Lactose phosphotransferase system repressor 34 0.27
GK12_HUMAN (P63130) HERV-K_1q22 provirus ancestral Gag polyprote... 34 0.36
GK11_HUMAN (P63145) HERV-K_22q11.21 provirus ancestral Gag polyp... 34 0.36
GK10_HUMAN (P87889) HERV-K_5q33.3 provirus ancestral Gag polypro... 34 0.36
GAK6_HUMAN (P62685) HERV-K_8p23.1 provirus ancestral Gag polypro... 34 0.36
GAK5_HUMAN (P62684) HERV-K_19p13.11 provirus ancestral Gag polyp... 34 0.36
GAK4_HUMAN (P63126) HERV-K_6q14.1 provirus ancestral Gag polypro... 34 0.36
GAK3_HUMAN (Q9YNA8) HERV-K_19q12 provirus ancestral Gag polyprot... 34 0.36
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 139 bits (351), Expect = 5e-33
Identities = 80/223 (35%), Positives = 124/223 (54%), Gaps = 11/223 (4%)
Query: 1 MSQAEKTEMVDKARSAIVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQ 60
M + ++ ++A SAI L L D V+ + E TA +W +LESLYM+K+L ++ +LK+Q
Sbjct: 47 MKAEDWADLDERAASAIRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQ 106
Query: 61 LYSFRMVESKAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKNTMLYG 120
LY+ M E + L FN ++ L N+ V++E+EDKAILLL +LP S+++ T+L+G
Sbjct: 107 LYALHMSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAILLLNSLPSSYDNLATTILHG 166
Query: 121 KEGTVTLEEIQAALRTKELTNSKD-------LTHEHDEGLSVSRGNGGGRGNRRKSGNKS 173
K T+ L+++ +AL E K +T S N G G R KS N+S
Sbjct: 167 KT-TIELKDVTSALLLNEKMRKKPENQGQALITEGRGRSYQRSSNNYGRSGARGKSKNRS 225
Query: 174 RF---ECFNCHKMGHFKKDCPEINGNSAQIVYEGYEDVGALMV 213
+ C+NC++ GHFK+DCP + + +D A MV
Sbjct: 226 KSRVRNCYNCNQPGHFKRDCPNPRKGKGETSGQKNDDNTAAMV 268
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 65.9 bits (159), Expect = 8e-11
Identities = 46/191 (24%), Positives = 87/191 (45%), Gaps = 13/191 (6%)
Query: 13 ARSAIVLCLGDKVLREVAKEATAASMWAKLESLYMTKSLAHRQFLKQQLYSFRMVESKAI 72
A+S I+ L D L + TA + L+++Y KSLA + L+++L S ++ ++
Sbjct: 55 AKSTIIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSL 114
Query: 73 MEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKNTMLYGKEGTVTLEEIQA 132
+ F++++ +L ++E+ DK LL LP ++ + E +TL ++
Sbjct: 115 LSHFHIFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSEENLTLAFVKN 174
Query: 133 ALRTKELT--NSKDLTHEHDEGLSVSRGNGGGRGNRRKS-----------GNKSRFECFN 179
L +E+ N + T + V N + N K+ +K + +C +
Sbjct: 175 RLLDQEIKIKNDHNDTSKKVMNAIVHNNNNTYKNNLFKNRVTKPKKIFKGNSKYKVKCHH 234
Query: 180 CHKMGHFKKDC 190
C + GH KKDC
Sbjct: 235 CGREGHIKKDC 245
>HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding
protein)
Length = 271
Score = 37.4 bits (85), Expect = 0.032
Identities = 24/81 (29%), Positives = 33/81 (40%), Gaps = 18/81 (22%)
Query: 158 GNGGGRGNRR-KSGNKSRFE----CFNCHKMGHFKKDCPEINGNSAQIVYEGYEDVGALM 212
G+ GG G +R +SG + + C+ C GH +DCP G GY G
Sbjct: 118 GSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQG--------GYSGAGDRT 169
Query: 213 VWCCLEDEEGDVSHLGSDACN 233
+ C GD H+ D N
Sbjct: 170 CYKC-----GDAGHISRDCPN 185
Score = 37.0 bits (84), Expect = 0.042
Identities = 14/33 (42%), Positives = 17/33 (51%)
Query: 165 NRRKSGNKSRFECFNCHKMGHFKKDCPEINGNS 197
N K G FEC+ C + GH +DCP G S
Sbjct: 87 NSAKPGAAKGFECYKCGQEGHLSRDCPSSQGGS 119
Score = 33.1 bits (74), Expect = 0.61
Identities = 14/42 (33%), Positives = 19/42 (44%)
Query: 165 NRRKSGNKSRFECFNCHKMGHFKKDCPEINGNSAQIVYEGYE 206
N +SG CF C + GH +DCP A +E Y+
Sbjct: 60 NEARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKGFECYK 101
Score = 32.3 bits (72), Expect = 1.0
Identities = 26/92 (28%), Positives = 37/92 (39%), Gaps = 29/92 (31%)
Query: 158 GNGG--GRGNRR-----KSGNKSRF------------ECFNCHKMGHFKKDCPEINGNSA 198
G GG G G+R+ +SG+ SR C+ C K GH ++CPE G+
Sbjct: 186 GQGGYSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRACYKCGKPGHISRECPEAGGS-- 243
Query: 199 QIVYEGYEDVGALMVWCCLEDEEGDVSHLGSD 230
Y G G + C G+ H+ D
Sbjct: 244 ---YGGSRGGGDRTCYKC-----GEAGHISRD 267
Score = 32.0 bits (71), Expect = 1.4
Identities = 12/32 (37%), Positives = 18/32 (55%), Gaps = 3/32 (9%)
Query: 160 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCP 191
GG G R G+++ C+ C + GH +DCP
Sbjct: 241 GGSYGGSRGGGDRT---CYKCGEAGHISRDCP 269
Score = 31.6 bits (70), Expect = 1.8
Identities = 18/64 (28%), Positives = 27/64 (42%), Gaps = 11/64 (17%)
Query: 155 VSRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGNSAQIVYEGYEDVGALMVW 214
+SR G+G +G+++ C+ C GH +DCP G GY G +
Sbjct: 151 ISRDCPNGQGGYSGAGDRT---CYKCGDAGHISRDCPNGQG--------GYSGAGDRKCY 199
Query: 215 CCLE 218
C E
Sbjct: 200 KCGE 203
Score = 30.0 bits (66), Expect = 5.1
Identities = 10/26 (38%), Positives = 16/26 (61%)
Query: 167 RKSGNKSRFECFNCHKMGHFKKDCPE 192
++ +S C NC K GH+ ++CPE
Sbjct: 8 KRPRTESSTSCRNCGKEGHYARECPE 33
Score = 29.3 bits (64), Expect = 8.8
Identities = 16/65 (24%), Positives = 27/65 (40%), Gaps = 14/65 (21%)
Query: 170 GNKSRFECFNCHKMGHFKKDCPEINGNSAQIVYEGYEDVGALMVWCCLEDEEGDVSHLGS 229
G++ CF C + GH ++CP + A GA+ + C G+ H+
Sbjct: 38 GDERSTTCFRCGEEGHMSRECPNEARSGA---------AGAMTCFRC-----GEAGHMSR 83
Query: 230 DACNT 234
D N+
Sbjct: 84 DCPNS 88
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 37.0 bits (84), Expect = 0.042
Identities = 18/41 (43%), Positives = 22/41 (52%), Gaps = 4/41 (9%)
Query: 152 GLSVSRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPE 192
G S G+GGG +R + N CFNC + GH DCPE
Sbjct: 352 GNSNGFGSGGGGQDRGERNNN----CFNCQQPGHRSNDCPE 388
Score = 37.0 bits (84), Expect = 0.042
Identities = 23/73 (31%), Positives = 28/73 (37%), Gaps = 19/73 (26%)
Query: 139 LTNSKDLTHEHDEGLSVSRGNGGGRGNRRKS-----GNKSRF--------------ECFN 179
LTN + H+ G + G G GN S GN + F CFN
Sbjct: 202 LTNGFGSGNNHESGFGGGKSGGFGGGNSGGSGFGSGGNSNGFGSGGGGQDRGERNNNCFN 261
Query: 180 CHKMGHFKKDCPE 192
C + GH DCPE
Sbjct: 262 CQQPGHRSNDCPE 274
Score = 33.1 bits (74), Expect = 0.61
Identities = 15/35 (42%), Positives = 18/35 (50%), Gaps = 3/35 (8%)
Query: 158 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPE 192
GN G GN + G +CFNC GH +CPE
Sbjct: 439 GNAEGFGNNEERGP---MKCFNCKGEGHRSAECPE 470
Score = 30.8 bits (68), Expect = 3.0
Identities = 21/69 (30%), Positives = 31/69 (44%), Gaps = 5/69 (7%)
Query: 159 NGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGNSAQIVYEGYEDVGALMVWCCLE 218
N G G+R + CFNC + GH +CP N A+ EG E G + +E
Sbjct: 457 NCKGEGHRSAECPEPPRGCFNCGEQGHRSNECP----NPAK-PREGAEGEGPKATYVPVE 511
Query: 219 DEEGDVSHL 227
D +V ++
Sbjct: 512 DNMEEVFNM 520
Score = 30.0 bits (66), Expect = 5.1
Identities = 9/16 (56%), Positives = 12/16 (74%)
Query: 177 CFNCHKMGHFKKDCPE 192
C+NC + GH +DCPE
Sbjct: 284 CYNCQQPGHNSRDCPE 299
Score = 30.0 bits (66), Expect = 5.1
Identities = 9/16 (56%), Positives = 12/16 (74%)
Query: 177 CFNCHKMGHFKKDCPE 192
C+NC + GH +DCPE
Sbjct: 398 CYNCQQPGHNSRDCPE 413
>GAG_MMTVC (P11284) Gag polyprotein [Contains: Protein p10;
Phosphorylated protein pp21; Protein p3; Protein p8;
Major core protein p27; Nucleic acid binding protein
p14]
Length = 591
Score = 37.0 bits (84), Expect = 0.042
Identities = 17/42 (40%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query: 155 VSRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGN 196
V + GGG+G + G CF+C K GH K+DC E G+
Sbjct: 509 VKQTYGGGKGGQGSKGPV----CFSCGKTGHIKRDCKEEKGS 546
>GAG_IPMA (P11365) Retrovirus-related Gag polyprotein [Contains:
Protease (EC 3.4.23.-)]
Length = 827
Score = 37.0 bits (84), Expect = 0.042
Identities = 17/26 (65%), Positives = 17/26 (65%), Gaps = 1/26 (3%)
Query: 165 NRRKSGNKSRFECFNCHKMGHFKKDC 190
NR S N R CFNC K GHFKKDC
Sbjct: 449 NRSMSRNDQR-TCFNCGKPGHFKKDC 473
>GAG_SIVA1 (P27972) Gag polyprotein [Contains: Core protein p17;
Core protein p24; Core protein p15]
Length = 520
Score = 35.8 bits (81), Expect = 0.094
Identities = 15/33 (45%), Positives = 19/33 (57%), Gaps = 5/33 (15%)
Query: 160 GGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPE 192
GGGRG R +C+NC K GH ++ CPE
Sbjct: 388 GGGRGRPRPPP-----KCYNCGKFGHMQRQCPE 415
>Y893_DROME (Q9VV43) Hypothetical protein CG4893
Length = 192
Score = 35.4 bits (80), Expect = 0.12
Identities = 32/113 (28%), Positives = 44/113 (38%), Gaps = 16/113 (14%)
Query: 70 KAIMEQLTEFNKILDDLENIEVQLEDEDKAILLLCALPKSFESFKNTMLYGKEGTVTLEE 129
KA+ L+++NK LDDL + E K L C P G V++
Sbjct: 89 KAMKISLSDYNKFLDDLAKTKKVELSEIKQKLASCGAP---------------GVVSVSA 133
Query: 130 IQAALRTKELTNSKDLTHEHDEGLSVSRGNGGGRGNRRKSGNKSRFECFNCHK 182
+AA LT++ T H E S G G G RR + S + HK
Sbjct: 134 GKAAAAVDRLTDTSKYTGSHKERFDAS-GKGKGIAGRRNVVDGSGYVSGYQHK 185
>GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-)
(Germline helicase-1)
Length = 763
Score = 35.4 bits (80), Expect = 0.12
Identities = 14/35 (40%), Positives = 16/35 (45%)
Query: 158 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPE 192
G G G G + CFNC + GH DCPE
Sbjct: 141 GGNSGFGEGGHGGGERNNNCFNCQQPGHRSSDCPE 175
Score = 32.7 bits (73), Expect = 0.79
Identities = 22/69 (31%), Positives = 31/69 (44%), Gaps = 5/69 (7%)
Query: 159 NGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGNSAQIVYEGYEDVGALMVWCCLE 218
N G G+R + CFNC + GH +CP N A+ EG E G + +E
Sbjct: 246 NCKGEGHRSAECPEPPRGCFNCGEQGHRSNECP----NPAK-PREGVEGEGPKATYVPVE 300
Query: 219 DEEGDVSHL 227
D DV ++
Sbjct: 301 DNMEDVFNM 309
Score = 32.7 bits (73), Expect = 0.79
Identities = 19/65 (29%), Positives = 25/65 (38%), Gaps = 14/65 (21%)
Query: 142 SKDLTHEHDEGLSVSRGNGGGRGNRRKSGN--------------KSRFECFNCHKMGHFK 187
S++ T E + G GGG G GN + +CFNC GH
Sbjct: 195 SRECTEERKPREGRTGGFGGGAGFGNNGGNDGFGGDGGFGGGEERGPMKCFNCKGEGHRS 254
Query: 188 KDCPE 192
+CPE
Sbjct: 255 AECPE 259
>GAG_MMTVB (P10258) Gag polyprotein [Contains: Protein p10;
Phosphorylated protein pp21; Protein p3; Protein p8;
Major core protein p27; Nucleic acid binding protein
p14]
Length = 591
Score = 35.4 bits (80), Expect = 0.12
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query: 155 VSRGNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEINGN 196
V + GGG+G + G CF+C K GH +KDC + G+
Sbjct: 509 VKQTYGGGKGGQGAEGPV----CFSCGKTGHIRKDCKDEKGS 546
>GRP2_NICSY (P27484) Glycine-rich protein 2
Length = 214
Score = 35.0 bits (79), Expect = 0.16
Identities = 15/38 (39%), Positives = 18/38 (46%), Gaps = 3/38 (7%)
Query: 158 GNGGGRGNRRKSGNKSRFECFNCHKMGHFKKDCPEING 195
G GGG G G CF C + GHF +DC + G
Sbjct: 143 GGGGGYGGGGSGGGSG---CFKCGESGHFARDCSQSGG 177
>GAG_SRV2 (P51516) Gag polyprotein (Core polyprotein) [Contains:
Core protein p10; Core phosphoprotein p18; Core protein
p12; Core protein p27; Core protein p14; Core protein
p4]
Length = 654
Score = 34.7 bits (78), Expect = 0.21
Identities = 17/48 (35%), Positives = 25/48 (51%), Gaps = 1/48 (2%)
Query: 144 DLTHEHDEGLSVSRGNGGGR-GNRRKSGNKSRFECFNCHKMGHFKKDC 190
D+ + +GL+++ G + + NK R CF C K GHF KDC
Sbjct: 512 DIGPSYQQGLAMAAAFSGQTVKDLLNNKNKDRGGCFKCGKKGHFAKDC 559
>LACR_STRMU (P26422) Lactose phosphotransferase system repressor
Length = 251
Score = 34.3 bits (77), Expect = 0.27
Identities = 29/96 (30%), Positives = 44/96 (45%), Gaps = 19/96 (19%)
Query: 116 TMLYGKEGTVTLEEIQAALRTKELTNSKDLTHEHDEGLSV-SRGNGGGRGNRRKSGNKSR 174
T L K GT+ + E+ L+ ++T +DLT EGL V +R +GG R N
Sbjct: 11 TKLINKRGTIRVTEVVERLKVSDMTVRRDLTEL--EGLGVLTRIHGGARSN--------- 59
Query: 175 FECFNCHKMGHFKKDCPEIN------GNSAQIVYEG 204
F +M H +K +I +A++V EG
Sbjct: 60 -NIFQYKEMSHEEKHSRQIEEKHYIAQKAAELVEEG 94
>GK12_HUMAN (P63130) HERV-K_1q22 provirus ancestral Gag polyprotein
(Gag polyprotein) (HERV-K102 Gag protein) (HERV-K(III)
Gag protein) [Contains: Matrix protein; Capsid protein;
Nucleocapsid protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.36
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 176 ECFNCHKMGHFKKDCPEINGNSAQI 200
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GK11_HUMAN (P63145) HERV-K_22q11.21 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K101 Gag protein)
[Contains: Matrix protein; Capsid protein; Nucleocapsid
protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.36
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 176 ECFNCHKMGHFKKDCPEINGNSAQI 200
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GK10_HUMAN (P87889) HERV-K_5q33.3 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K10 Gag protein)
(HERV-K107 Gag protein) [Contains: Matrix protein;
Capsid protein; Nucleocapsid protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.36
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 176 ECFNCHKMGHFKKDCPEINGNSAQI 200
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GAK6_HUMAN (P62685) HERV-K_8p23.1 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K115 Gag protein)
[Contains: Matrix protein; Capsid protein; Nucleocapsid
protein]
Length = 646
Score = 33.9 bits (76), Expect = 0.36
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 176 ECFNCHKMGHFKKDCPEINGNSAQI 200
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GAK5_HUMAN (P62684) HERV-K_19p13.11 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K113 Gag protein)
[Contains: Matrix protein; Capsid protein; Nucleocapsid
protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.36
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 176 ECFNCHKMGHFKKDCPEINGNSAQI 200
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GAK4_HUMAN (P63126) HERV-K_6q14.1 provirus ancestral Gag
polyprotein (Gag polyprotein) (HERV-K109 Gag protein)
(HERV-K(C6) Gag protein) [Contains: Matrix protein;
Capsid protein; Nucleocapsid protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.36
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 176 ECFNCHKMGHFKKDCPEINGNSAQI 200
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
>GAK3_HUMAN (Q9YNA8) HERV-K_19q12 provirus ancestral Gag polyprotein
(Gag polyprotein) (HERV-K(C19) Gag protein) [Contains:
Matrix protein; Capsid protein; Nucleocapsid protein]
Length = 665
Score = 33.9 bits (76), Expect = 0.36
Identities = 11/25 (44%), Positives = 18/25 (72%)
Query: 176 ECFNCHKMGHFKKDCPEINGNSAQI 200
+C+NC ++GH KK+CP +N + I
Sbjct: 544 KCYNCGQIGHLKKNCPVLNKQNITI 568
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.132 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,044,173
Number of Sequences: 164201
Number of extensions: 1117014
Number of successful extensions: 4437
Number of sequences better than 10.0: 95
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 4283
Number of HSP's gapped (non-prelim): 175
length of query: 240
length of database: 59,974,054
effective HSP length: 107
effective length of query: 133
effective length of database: 42,404,547
effective search space: 5639804751
effective search space used: 5639804751
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC146972.3


