

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146941.10 - phase: 0
(124 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
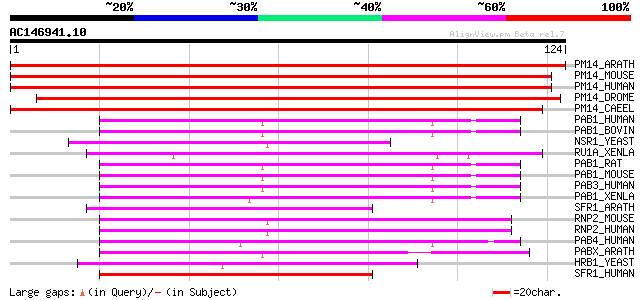
Score E
Sequences producing significant alignments: (bits) Value
PM14_ARATH (Q9FMP4) Pre-mRNA branch site p14-like protein 229 8e-61
PM14_MOUSE (P59708) Pre-mRNA branch site protein p14 (SF3B 14 kD... 171 2e-43
PM14_HUMAN (Q9Y3B4) Pre-mRNA branch site protein p14 (SF3B 14 kD... 171 2e-43
PM14_DROME (Q9VRV7) Pre-mRNA branch site p14-like protein 166 7e-42
PM14_CAEEL (Q8ITY4) Pre-mRNA branch site p14-like protein 157 3e-39
PAB1_HUMAN (P11940) Polyadenylate-binding protein 1 (Poly(A)-bin... 52 3e-07
PAB1_BOVIN (P61286) Polyadenylate-binding protein 1 (Poly(A)-bin... 52 3e-07
NSR1_YEAST (P27476) Nuclear localization sequence binding protei... 51 4e-07
RU1A_XENLA (P45429) U1 small nuclear ribonucleoprotein A (U1 snR... 50 9e-07
PAB1_RAT (Q9EPH8) Polyadenylate-binding protein 1 (Poly(A)-bindi... 50 9e-07
PAB1_MOUSE (P29341) Polyadenylate-binding protein 1 (Poly(A)-bin... 50 9e-07
PAB3_HUMAN (Q9H361) Polyadenylate-binding protein 3 (Poly(A)-bin... 49 2e-06
PAB1_XENLA (P20965) Polyadenylate-binding protein 1 (Poly(A)-bin... 49 2e-06
SFR1_ARATH (O22315) Pre-mRNA splicing factor SF2 (SR1 protein) 49 3e-06
RNP2_MOUSE (Q8VH51) RNA-binding region containing protein 2 (Coa... 49 3e-06
RNP2_HUMAN (Q14498) RNA-binding region containing protein 2 (Hep... 49 3e-06
PAB4_HUMAN (Q13310) Polyadenylate-binding protein 4 (Poly(A)-bin... 49 3e-06
PABX_ARATH (Q9ZQA8) Probable polyadenylate-binding protein At2g3... 48 4e-06
HRB1_YEAST (P38922) HRB1 protein (TOM34 protein) 48 4e-06
SFR1_HUMAN (Q07955) Splicing factor, arginine/serine-rich 1 (pre... 47 6e-06
>PM14_ARATH (Q9FMP4) Pre-mRNA branch site p14-like protein
Length = 124
Score = 229 bits (584), Expect = 8e-61
Identities = 113/124 (91%), Positives = 117/124 (94%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
MTTISLRK NTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIG +K T+GTA
Sbjct: 1 MTTISLRKSNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGCDKATKGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
FVVYEDIYDAK AVDHLSGFNVANRYLIVLYYQ AKMSKKFDQKK EDEI ++QEKYGVS
Sbjct: 61 FVVYEDIYDAKNAVDHLSGFNVANRYLIVLYYQHAKMSKKFDQKKSEDEITKLQEKYGVS 120
Query: 121 TKDK 124
TKDK
Sbjct: 121 TKDK 124
>PM14_MOUSE (P59708) Pre-mRNA branch site protein p14 (SF3B 14 kDa
subunit)
Length = 125
Score = 171 bits (434), Expect = 2e-43
Identities = 77/121 (63%), Positives = 98/121 (80%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + ++ N RLPPEVNR+LY+RNLP+ IT+EEMYDIFGKYG IRQIR+G +TRGTA
Sbjct: 1 MAMQAAKRANIRLPPEVNRILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGNTPETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
+VVYEDI+DAK A DHLSGFNV NRYL+VLYY + +K D KKKE+++ ++EKYG++
Sbjct: 61 YVVYEDIFDAKNACDHLSGFNVCNRYLVVLYYNANRAFQKMDTKKKEEQLKLLKEKYGIN 120
Query: 121 T 121
T
Sbjct: 121 T 121
>PM14_HUMAN (Q9Y3B4) Pre-mRNA branch site protein p14 (SF3B 14 kDa
subunit) (CGI-110) (HSPC175) (Ht006)
Length = 125
Score = 171 bits (434), Expect = 2e-43
Identities = 77/121 (63%), Positives = 98/121 (80%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + ++ N RLPPEVNR+LY+RNLP+ IT+EEMYDIFGKYG IRQIR+G +TRGTA
Sbjct: 1 MAMQAAKRANIRLPPEVNRILYIRNLPYKITAEEMYDIFGKYGPIRQIRVGNTPETRGTA 60
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVS 120
+VVYEDI+DAK A DHLSGFNV NRYL+VLYY + +K D KKKE+++ ++EKYG++
Sbjct: 61 YVVYEDIFDAKNACDHLSGFNVCNRYLVVLYYNANRAFQKMDTKKKEEQLKLLKEKYGIN 120
Query: 121 T 121
T
Sbjct: 121 T 121
>PM14_DROME (Q9VRV7) Pre-mRNA branch site p14-like protein
Length = 121
Score = 166 bits (421), Expect = 7e-42
Identities = 76/117 (64%), Positives = 95/117 (80%)
Query: 7 RKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYED 66
++ + RLPPEVNR+LYVRNLP+ ITS+EMYDIFGK+GAIRQIR+G +TRGTAFVVYED
Sbjct: 3 KRNHIRLPPEVNRLLYVRNLPYKITSDEMYDIFGKFGAIRQIRVGNTPETRGTAFVVYED 62
Query: 67 IYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGVSTKD 123
I+DAK A DHLSGFNV NRYL+VLYYQ K K+ D KK++E+ ++ KY + T +
Sbjct: 63 IFDAKNACDHLSGFNVCNRYLVVLYYQSNKAFKRVDMDKKQEELNNIKAKYNLKTPE 119
>PM14_CAEEL (Q8ITY4) Pre-mRNA branch site p14-like protein
Length = 188
Score = 157 bits (398), Expect = 3e-39
Identities = 69/119 (57%), Positives = 95/119 (78%)
Query: 1 MTTISLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTA 60
M + + +LPPEVNR+LY++NLP+ IT+EEMY+IFGK+GA+RQIR+G +TRGTA
Sbjct: 51 MAMANRQNRGAKLPPEVNRILYIKNLPYKITTEEMYEIFGKFGAVRQIRVGNTAETRGTA 110
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYGV 119
FVVYEDI+DAKTA +HLSG+NV+NRYL+VLYYQ K K+ D +K ++ ++E+YG+
Sbjct: 111 FVVYEDIFDAKTACEHLSGYNVSNRYLVVLYYQATKAWKRMDTEKARTKLEDIKERYGI 169
>PAB1_HUMAN (P11940) Polyadenylate-binding protein 1
(Poly(A)-binding protein 1) (PABP 1)
Length = 636
Score = 51.6 bits (122), Expect = 3e-07
Identities = 30/101 (29%), Positives = 57/101 (55%), Gaps = 8/101 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
+Y++N ++ E + D+FGK+G +++ T++ ++G FV +E DA+ AVD ++
Sbjct: 193 VYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMN 252
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G + + + V Q Q ++ +KF+Q K+D I R Q
Sbjct: 253 GKELNGKQIYVGRAQKKVERQTELKRKFEQ-MKQDRITRYQ 292
Score = 36.2 bits (82), Expect = 0.013
Identities = 18/77 (23%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
++++NL +I ++ +YD F +G I ++ +++ ++G FV +E A+ A++ ++G
Sbjct: 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNG 160
Query: 80 FNVANRYLIVLYYQQAK 96
+ +R + V ++ K
Sbjct: 161 MLLNDRKVFVGRFKSRK 177
Score = 33.5 bits (75), Expect = 0.086
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 6/90 (6%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
LYV+NL I E + F +G I ++ ++G FV + +A AV ++G
Sbjct: 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query: 80 FNVANRYLIVLYYQ-----QAKMSKKFDQK 104
VA + L V Q QA ++ ++ Q+
Sbjct: 356 RIVATKPLYVALAQRKEERQAHLTNQYMQR 385
Score = 32.7 bits (73), Expect = 0.15
Identities = 22/83 (26%), Positives = 44/83 (52%), Gaps = 3/83 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
LYV +L ++T +Y+ F G I IR+ + TR G A+V ++ DA+ A+D +
Sbjct: 13 LYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTM 72
Query: 78 SGFNVANRYLIVLYYQQAKMSKK 100
+ + + + +++ Q+ +K
Sbjct: 73 NFDVIKGKPVRIMWSQRDPSLRK 95
>PAB1_BOVIN (P61286) Polyadenylate-binding protein 1
(Poly(A)-binding protein 1) (PABP 1)
Length = 636
Score = 51.6 bits (122), Expect = 3e-07
Identities = 30/101 (29%), Positives = 57/101 (55%), Gaps = 8/101 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
+Y++N ++ E + D+FGK+G +++ T++ ++G FV +E DA+ AVD ++
Sbjct: 193 VYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMN 252
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G + + + V Q Q ++ +KF+Q K+D I R Q
Sbjct: 253 GKELNGKQIYVGRAQKKVERQTELKRKFEQ-MKQDRITRYQ 292
Score = 36.2 bits (82), Expect = 0.013
Identities = 18/77 (23%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
++++NL +I ++ +YD F +G I ++ +++ ++G FV +E A+ A++ ++G
Sbjct: 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNG 160
Query: 80 FNVANRYLIVLYYQQAK 96
+ +R + V ++ K
Sbjct: 161 MLLNDRKVFVGRFKSRK 177
Score = 33.5 bits (75), Expect = 0.086
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 6/90 (6%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
LYV+NL I E + F +G I ++ ++G FV + +A AV ++G
Sbjct: 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query: 80 FNVANRYLIVLYYQ-----QAKMSKKFDQK 104
VA + L V Q QA ++ ++ Q+
Sbjct: 356 RIVATKPLYVALAQRKEERQAHLTNQYMQR 385
Score = 32.7 bits (73), Expect = 0.15
Identities = 22/83 (26%), Positives = 44/83 (52%), Gaps = 3/83 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
LYV +L ++T +Y+ F G I IR+ + TR G A+V ++ DA+ A+D +
Sbjct: 13 LYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTM 72
Query: 78 SGFNVANRYLIVLYYQQAKMSKK 100
+ + + + +++ Q+ +K
Sbjct: 73 NFDVIKGKPVRIMWSQRDPSLRK 95
>NSR1_YEAST (P27476) Nuclear localization sequence binding protein
(P67)
Length = 414
Score = 51.2 bits (121), Expect = 4e-07
Identities = 25/75 (33%), Positives = 45/75 (59%), Gaps = 3/75 (4%)
Query: 14 PPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDA 70
P E + L++ NL FN + ++++F K+G + +RI T+ +T +G +V + ++ DA
Sbjct: 262 PSEPSDTLFLGNLSFNADRDAIFELFAKHGEVVSVRIPTHPETEQPKGFGYVQFSNMEDA 321
Query: 71 KTAVDHLSGFNVANR 85
K A+D L G + NR
Sbjct: 322 KKALDALQGEYIDNR 336
>RU1A_XENLA (P45429) U1 small nuclear ribonucleoprotein A (U1 snRNP
protein A) (U1A protein) (U1-A)
Length = 282
Score = 50.1 bits (118), Expect = 9e-07
Identities = 33/114 (28%), Positives = 56/114 (48%), Gaps = 12/114 (10%)
Query: 18 NRVLYVRNLPFNITSEEM----YDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTA 73
N +Y+ NL I +E+ Y IF ++G I I + N RG AFV++++ A A
Sbjct: 9 NNTIYINNLNEKIKKDELKKSLYAIFSQFGQILDILVSRNLKMRGQAFVIFKETSSATNA 68
Query: 74 VDHLSGFNVANRYLIVLYYQQ-----AKMSKKF---DQKKKEDEIARMQEKYGV 119
+ + GF ++ + + Y + AKM F D+K++E ++ E GV
Sbjct: 69 LRSMQGFPFYDKPMRIQYSKTDSDIIAKMKGTFVERDRKRQEKRKVKVPEVQGV 122
>PAB1_RAT (Q9EPH8) Polyadenylate-binding protein 1 (Poly(A)-binding
protein 1) (PABP 1)
Length = 636
Score = 50.1 bits (118), Expect = 9e-07
Identities = 29/101 (28%), Positives = 57/101 (55%), Gaps = 8/101 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
+Y++N ++ E + ++FGK+G +++ T++ ++G FV +E DA+ AVD ++
Sbjct: 193 VYIKNFGEDMDDERLKELFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMN 252
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G + + + V Q Q ++ +KF+Q K+D I R Q
Sbjct: 253 GKELNGKQIYVGRAQKKVERQTELKRKFEQ-MKQDRITRYQ 292
Score = 36.2 bits (82), Expect = 0.013
Identities = 18/77 (23%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
++++NL +I ++ +YD F +G I ++ +++ ++G FV +E A+ A++ ++G
Sbjct: 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNG 160
Query: 80 FNVANRYLIVLYYQQAK 96
+ +R + V ++ K
Sbjct: 161 MLLNDRKVFVGRFKSRK 177
Score = 33.5 bits (75), Expect = 0.086
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 6/90 (6%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
LYV+NL I E + F +G I ++ ++G FV + +A AV ++G
Sbjct: 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query: 80 FNVANRYLIVLYYQ-----QAKMSKKFDQK 104
VA + L V Q QA ++ ++ Q+
Sbjct: 356 RIVATKPLYVALAQRKEERQAHLTNQYMQR 385
Score = 32.7 bits (73), Expect = 0.15
Identities = 22/83 (26%), Positives = 44/83 (52%), Gaps = 3/83 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
LYV +L ++T +Y+ F G I IR+ + TR G A+V ++ DA+ A+D +
Sbjct: 13 LYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTM 72
Query: 78 SGFNVANRYLIVLYYQQAKMSKK 100
+ + + + +++ Q+ +K
Sbjct: 73 NFDVIKGKPVRIMWSQRDPSLRK 95
>PAB1_MOUSE (P29341) Polyadenylate-binding protein 1
(Poly(A)-binding protein 1) (PABP 1)
Length = 636
Score = 50.1 bits (118), Expect = 9e-07
Identities = 29/101 (28%), Positives = 57/101 (55%), Gaps = 8/101 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
+Y++N ++ E + ++FGK+G +++ T++ ++G FV +E DA+ AVD ++
Sbjct: 193 VYIKNFGEDMDDERLKELFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMN 252
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G + + + V Q Q ++ +KF+Q K+D I R Q
Sbjct: 253 GKELNGKQIYVGRAQKKVERQTELKRKFEQ-MKQDRITRYQ 292
Score = 37.7 bits (86), Expect = 0.005
Identities = 26/105 (24%), Positives = 58/105 (54%), Gaps = 10/105 (9%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
++++NL +I ++ +YD F +G I ++ +++ ++G FV +E A+ A++ ++G
Sbjct: 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYGFVHFETQEAAERAIEKMNG 160
Query: 80 FNVANRYLIVLYYQQAKMSKKFDQKKKEDEI-ARMQEKYGVSTKD 123
+ +R + V ++ QK++E E+ AR +E V K+
Sbjct: 161 MLLNDRKVFVGRFK--------SQKEREAELGARAKEFTNVYIKN 197
Score = 33.5 bits (75), Expect = 0.086
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 6/90 (6%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
LYV+NL I E + F +G I ++ ++G FV + +A AV ++G
Sbjct: 296 LYVKNLDDGIDDERLRKEFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query: 80 FNVANRYLIVLYYQ-----QAKMSKKFDQK 104
VA + L V Q QA ++ ++ Q+
Sbjct: 356 RIVATKPLYVALAQRKEERQAHLTNQYMQR 385
Score = 32.7 bits (73), Expect = 0.15
Identities = 22/83 (26%), Positives = 44/83 (52%), Gaps = 3/83 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
LYV +L ++T +Y+ F G I IR+ + TR G A+V ++ DA+ A+D +
Sbjct: 13 LYVGDLHPDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTM 72
Query: 78 SGFNVANRYLIVLYYQQAKMSKK 100
+ + + + +++ Q+ +K
Sbjct: 73 NFDVIKGKPVRIMWSQRDPSLRK 95
>PAB3_HUMAN (Q9H361) Polyadenylate-binding protein 3
(Poly(A)-binding protein 3) (PABP 3) (Testis-specific
poly(A)-binding protein)
Length = 631
Score = 49.3 bits (116), Expect = 2e-06
Identities = 29/101 (28%), Positives = 56/101 (54%), Gaps = 8/101 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
+Y++N ++ E + D+FGK+G +++ T++ ++G FV +E DA+ AVD ++
Sbjct: 193 VYIKNFGEDMDDERLKDLFGKFGPALSVKVMTDESGKSKGFGFVSFERHEDAQKAVDEMN 252
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G + + + V Q Q ++ + F+Q K+D I R Q
Sbjct: 253 GKELNGKQIYVGRAQKKVERQTELKRTFEQ-MKQDRITRYQ 292
Score = 33.9 bits (76), Expect = 0.066
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 6/90 (6%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
LYV+NL I E + F +G I ++ ++G FV + +A AV ++G
Sbjct: 296 LYVKNLDDGIDDERLRKAFSPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query: 80 FNVANRYLIVLYYQ-----QAKMSKKFDQK 104
VA + L V Q QA ++ ++ Q+
Sbjct: 356 RIVATKPLYVALAQRKEERQAYLTNEYMQR 385
Score = 32.3 bits (72), Expect = 0.19
Identities = 18/77 (23%), Positives = 41/77 (52%), Gaps = 1/77 (1%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
++V+NL +I ++ +YD +G I + +++ ++G FV +E A+ A+ ++G
Sbjct: 101 IFVKNLDKSINNKALYDTVSAFGNILSCNVVCDENGSKGYGFVHFETHEAAERAIKKMNG 160
Query: 80 FNVANRYLIVLYYQQAK 96
+ R + V ++ K
Sbjct: 161 MLLNGRKVFVGQFKSRK 177
Score = 32.3 bits (72), Expect = 0.19
Identities = 24/94 (25%), Positives = 46/94 (48%), Gaps = 3/94 (3%)
Query: 10 NTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGT---AFVVYED 66
N P LYV +L ++T +Y+ F G I IRI + T G+ A+V ++
Sbjct: 2 NPSTPSYPTASLYVGDLHPDVTEAMLYEKFSPAGPILSIRICRDLITSGSSNYAYVNFQH 61
Query: 67 IYDAKTAVDHLSGFNVANRYLIVLYYQQAKMSKK 100
DA+ A+D ++ + + + +++ Q+ +K
Sbjct: 62 TKDAEHALDTMNFDVIKGKPVRIMWSQRDPSLRK 95
>PAB1_XENLA (P20965) Polyadenylate-binding protein 1
(Poly(A)-binding protein 1) (PABP 1)
Length = 633
Score = 49.3 bits (116), Expect = 2e-06
Identities = 30/101 (29%), Positives = 55/101 (53%), Gaps = 8/101 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGT--NKDTRGTAFVVYEDIYDAKTAVDHLS 78
+Y++N ++ E + ++FGKYG +++ T N ++G FV +E DA+ AVD +
Sbjct: 193 VYIKNFGDDMNDERLKEMFGKYGPALSVKVMTDDNGKSKGFGFVSFERHEDAQKAVDEMY 252
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G ++ + + V Q Q ++ +KF+Q +D I R Q
Sbjct: 253 GKDMNGKSMFVGRAQKKVERQTELKRKFEQ-MNQDRITRYQ 292
Score = 38.1 bits (87), Expect = 0.003
Identities = 25/115 (21%), Positives = 54/115 (46%), Gaps = 17/115 (14%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
++++NL +I ++ +YD F +G I ++ +++ ++G FV +E A+ A+D ++G
Sbjct: 101 IFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYGFVHFETQEAAERAIDKMNG 160
Query: 80 FNVANRYLIVLYYQQAK----------------MSKKFDQKKKEDEIARMQEKYG 118
+ +R + V ++ K K F ++ + M KYG
Sbjct: 161 MLLNDRKVFVGRFKSRKEREAELGARAKEFTNVYIKNFGDDMNDERLKEMFGKYG 215
Score = 34.3 bits (77), Expect = 0.051
Identities = 23/83 (27%), Positives = 44/83 (52%), Gaps = 3/83 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
LYV +L ++T +Y+ F G I IR+ + TR G A+V ++ DA+ A+D +
Sbjct: 13 LYVGDLHQDVTEAMLYEKFSPAGPILSIRVCRDMITRRSLGYAYVNFQQPADAERALDTM 72
Query: 78 SGFNVANRYLIVLYYQQAKMSKK 100
+ + R + +++ Q+ +K
Sbjct: 73 NFDVIKGRPVRIMWSQRDPSLRK 95
Score = 32.0 bits (71), Expect = 0.25
Identities = 25/90 (27%), Positives = 42/90 (45%), Gaps = 6/90 (6%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
LYV+NL I E + F +G I ++ ++G FV + +A AV ++G
Sbjct: 296 LYVKNLDDGIDDERLRKEFLPFGTITSAKVMMEGGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query: 80 FNVANRYLIVLYYQ-----QAKMSKKFDQK 104
VA + L V Q QA ++ ++ Q+
Sbjct: 356 RIVATKPLYVALAQRKEERQAHLTNQYMQR 385
>SFR1_ARATH (O22315) Pre-mRNA splicing factor SF2 (SR1 protein)
Length = 303
Score = 48.5 bits (114), Expect = 3e-06
Identities = 24/64 (37%), Positives = 37/64 (57%)
Query: 18 NRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHL 77
+R +YV NLP +I E+ D+F KYG + QI + G AFV ++D DA+ A+
Sbjct: 6 SRTVYVGNLPGDIREREVEDLFSKYGPVVQIDLKVPPRPPGYAFVEFDDARDAEDAIHGR 65
Query: 78 SGFN 81
G++
Sbjct: 66 DGYD 69
>RNP2_MOUSE (Q8VH51) RNA-binding region containing protein 2
(Coactivator of activating protein-1 and estrogen
receptors) (Coactivator of AP-1 and ERs) (Transcription
coactivator CAPER)
Length = 530
Score = 48.5 bits (114), Expect = 3e-06
Identities = 29/95 (30%), Positives = 49/95 (51%), Gaps = 3/95 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
LYV +L FNIT + + IF +G I I++ + +T +G F+ + D AK A++ L
Sbjct: 252 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 311
Query: 78 SGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIAR 112
+GF +A R + V + + + DE+ R
Sbjct: 312 NGFELAGRPMKVGHVTERTDASSASSFLDSDELER 346
Score = 30.8 bits (68), Expect = 0.56
Identities = 24/96 (25%), Positives = 46/96 (47%), Gaps = 6/96 (6%)
Query: 6 LRKGNTRLPPEVN--RVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTA 60
+R+ L PE R ++ L I ++ + F G +R +R+ +++++R G A
Sbjct: 138 VREPIDNLTPEERDARTVFCMQLAARIRPRDLEEFFSTVGKVRDVRMISDRNSRRSRGIA 197
Query: 61 FVVYEDIYDAKTAVDHLSGFNVANRYLIVLYYQQAK 96
+V + D+ + A+ L+G V +IV Q K
Sbjct: 198 YVEFVDVSSVRLAIG-LTGQRVLGVPIIVQASQAEK 232
>RNP2_HUMAN (Q14498) RNA-binding region containing protein 2
(Hepatocellular carcinoma protein 1) (Splicing factor
HCC1)
Length = 530
Score = 48.5 bits (114), Expect = 3e-06
Identities = 29/95 (30%), Positives = 49/95 (51%), Gaps = 3/95 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDT---RGTAFVVYEDIYDAKTAVDHL 77
LYV +L FNIT + + IF +G I I++ + +T +G F+ + D AK A++ L
Sbjct: 252 LYVGSLHFNITEDMLRGIFEPFGRIESIQLMMDSETGRSKGYGFITFSDSECAKKALEQL 311
Query: 78 SGFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIAR 112
+GF +A R + V + + + DE+ R
Sbjct: 312 NGFELAGRPMKVGHVTERTDASSASSFLDSDELER 346
>PAB4_HUMAN (Q13310) Polyadenylate-binding protein 4
(Poly(A)-binding protein 4) (PABP 4) (Inducible
poly(A)-binding protein) (iPABP) (Activated-platelet
protein-1) (APP-1)
Length = 644
Score = 48.5 bits (114), Expect = 3e-06
Identities = 28/101 (27%), Positives = 55/101 (53%), Gaps = 8/101 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRI--GTNKDTRGTAFVVYEDIYDAKTAVDHLS 78
+Y++N + E + ++F ++G +++ N ++G FV YE DA AV+ ++
Sbjct: 193 VYIKNFGEEVDDESLKELFSQFGKTLSVKVMRDPNGKSKGFGFVSYEKHEDANKAVEEMN 252
Query: 79 GFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDEIARMQ 114
G ++ + + V Q QA++ +KF+Q K+E I+R Q
Sbjct: 253 GKEISGKIIFVGRAQKKVERQAELKRKFEQLKQE-RISRYQ 292
Score = 36.6 bits (83), Expect = 0.010
Identities = 19/77 (24%), Positives = 45/77 (57%), Gaps = 1/77 (1%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
++++NL +I ++ +YD F +G I ++ +++ ++G AFV +E A A++ ++G
Sbjct: 101 VFIKNLDKSIDNKALYDTFSAFGNILSCKVVCDENGSKGYAFVHFETQEAADKAIEKMNG 160
Query: 80 FNVANRYLIVLYYQQAK 96
+ +R + V ++ K
Sbjct: 161 MLLNDRKVFVGRFKSRK 177
Score = 33.5 bits (75), Expect = 0.086
Identities = 23/95 (24%), Positives = 44/95 (46%), Gaps = 1/95 (1%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD-TRGTAFVVYEDIYDAKTAVDHLSG 79
LY++NL I E++ F +G+I ++ ++G FV + +A AV ++G
Sbjct: 296 LYIKNLDDTIDDEKLRKEFSPFGSITSAKVMLEDGRSKGFGFVCFSSPEEATKAVTEMNG 355
Query: 80 FNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQ 114
V ++ L V Q+ + K + +A M+
Sbjct: 356 RIVGSKPLYVALAQRKEERKAHLTNQYMQRVAGMR 390
Score = 33.5 bits (75), Expect = 0.086
Identities = 21/83 (25%), Positives = 44/83 (52%), Gaps = 3/83 (3%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTR---GTAFVVYEDIYDAKTAVDHL 77
LYV +L ++T +Y+ F G + IR+ + TR G A+V ++ DA+ A+D +
Sbjct: 13 LYVGDLHSDVTEAMLYEKFSPAGPVLSIRVCRDMITRRSLGYAYVNFQQPADAERALDTM 72
Query: 78 SGFNVANRYLIVLYYQQAKMSKK 100
+ + + + +++ Q+ +K
Sbjct: 73 NFDVIKGKPIRIMWSQRDPSLRK 95
>PABX_ARATH (Q9ZQA8) Probable polyadenylate-binding protein
At2g36660 (Poly(A)-binding protein At2g36660) (PABP)
Length = 609
Score = 47.8 bits (112), Expect = 4e-06
Identities = 30/98 (30%), Positives = 53/98 (53%), Gaps = 7/98 (7%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFVVYEDIYDAKTAVDHLS 78
++V+NLP ++T+ + D+F K+G I ++ T +D +RG FV +E A A+ L+
Sbjct: 114 VFVKNLPESVTNAVLQDMFKKFGNIVSCKVATLEDGKSRGYGFVQFEQEDAAHAAIQTLN 173
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEK 116
VA++ + V K KK D+ K E++ + K
Sbjct: 174 STIVADKEIYV-----GKFMKKTDRVKPEEKYTNLYMK 206
Score = 40.0 bits (92), Expect = 0.001
Identities = 30/120 (25%), Positives = 63/120 (52%), Gaps = 8/120 (6%)
Query: 6 LRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGT--NKDTRGTAFVV 63
++K + P E LY++NL +++ + + + F ++G I + I N+ RG AFV
Sbjct: 188 MKKTDRVKPEEKYTNLYMKNLDADVSEDLLREKFAEFGKIVSLAIAKDENRLCRGYAFVN 247
Query: 64 YEDIYDAKTAVDHLSGFNVANRYLIVLYYQ-----QAKMSKKFDQKKKEDE-IARMQEKY 117
+++ DA+ A + ++G ++ L V Q + + ++F +K +E + IA++ Y
Sbjct: 248 FDNPEDARRAAETVNGTKFGSKCLYVGRAQKKAEREQLLREQFKEKHEEQKMIAKVSNIY 307
Score = 30.4 bits (67), Expect = 0.73
Identities = 25/100 (25%), Positives = 44/100 (44%), Gaps = 13/100 (13%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNK--DTRGTAFVVYEDIYDAKTAVDHLS 78
+YV+N+ +T EE+ F + G I ++ ++ ++G FV + +A AV
Sbjct: 306 IYVKNVNVAVTEEELRKHFSQCGTITSTKLMCDEKGKSKGFGFVCFSTPEEAIDAVKTFH 365
Query: 79 GFNVANRYLIVLYYQQAKMSKKFDQKKKEDEIARMQEKYG 118
G + L V Q KKED ++Q ++G
Sbjct: 366 GQMFHGKPLYVAIAQ-----------KKEDRKMQLQVQFG 394
>HRB1_YEAST (P38922) HRB1 protein (TOM34 protein)
Length = 429
Score = 47.8 bits (112), Expect = 4e-06
Identities = 23/78 (29%), Positives = 41/78 (52%), Gaps = 2/78 (2%)
Query: 16 EVNRVLYVRNLPFNITSEEMYDIFGKYGAIR--QIRIGTNKDTRGTAFVVYEDIYDAKTA 73
E NR++Y NLPF+ ++YD+F G + ++R + G A V Y+++ DA
Sbjct: 348 ERNRLIYCSNLPFSTAKSDLYDLFETIGKVNNAELRYDSKGAPTGIAVVEYDNVDDADVC 407
Query: 74 VDHLSGFNVANRYLIVLY 91
++ L+ +N L + Y
Sbjct: 408 IERLNNYNYGGCDLDISY 425
Score = 34.7 bits (78), Expect = 0.039
Identities = 22/87 (25%), Positives = 45/87 (51%), Gaps = 3/87 (3%)
Query: 5 SLRKGNTRLPPEVNRVLYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKD--TRGTAFV 62
+L +G R + + V+ V+NLP ++ + + DIF + G + + + D + G+ V
Sbjct: 223 ALDRGELRHNRKTHEVI-VKNLPASVNWQALKDIFKECGNVAHADVELDGDGVSTGSGTV 281
Query: 63 VYEDIYDAKTAVDHLSGFNVANRYLIV 89
+ DI D A++ +G+++ L V
Sbjct: 282 SFYDIKDLHRAIEKYNGYSIEGNVLDV 308
>SFR1_HUMAN (Q07955) Splicing factor, arginine/serine-rich 1
(pre-mRNA splicing factor SF2, P33 subunit)
(Alternative splicing factor ASF-1) (OK/SW-cl.3)
Length = 247
Score = 47.4 bits (111), Expect = 6e-06
Identities = 25/61 (40%), Positives = 38/61 (61%)
Query: 21 LYVRNLPFNITSEEMYDIFGKYGAIRQIRIGTNKDTRGTAFVVYEDIYDAKTAVDHLSGF 80
+YV NLP +I ++++ D+F KYGAIR I + + AFV +ED DA+ AV G+
Sbjct: 17 IYVGNLPPDIRTKDIEDVFYKYGAIRDIDLKNRRGGPPFAFVEFEDPRDAEDAVYGRDGY 76
Query: 81 N 81
+
Sbjct: 77 D 77
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,664,531
Number of Sequences: 164201
Number of extensions: 501904
Number of successful extensions: 1907
Number of sequences better than 10.0: 309
Number of HSP's better than 10.0 without gapping: 172
Number of HSP's successfully gapped in prelim test: 137
Number of HSP's that attempted gapping in prelim test: 1461
Number of HSP's gapped (non-prelim): 478
length of query: 124
length of database: 59,974,054
effective HSP length: 100
effective length of query: 24
effective length of database: 43,553,954
effective search space: 1045294896
effective search space used: 1045294896
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146941.10


