

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.6 - phase: 0
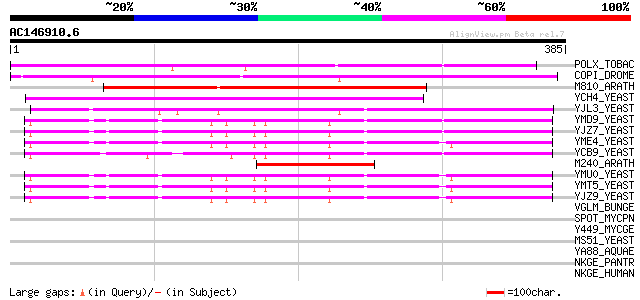
(385 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 228 3e-59
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 213 7e-55
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 196 1e-49
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 135 2e-31
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 105 1e-22
YMD9_YEAST (Q03434) Transposon Ty1 protein B 97 7e-20
YJZ7_YEAST (P47098) Transposon Ty1 protein B 96 2e-19
YME4_YEAST (Q04711) Transposon Ty1 protein B 94 4e-19
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 94 8e-19
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 90 1e-17
YMU0_YEAST (Q04670) Transposon Ty1 protein B 89 2e-17
YMT5_YEAST (Q04214) Transposon Ty1 protein B 87 7e-17
YJZ9_YEAST (P47100) Transposon Ty1 protein B 87 9e-17
VGLM_BUNGE (P12430) M polyprotein precursor [Contains: Glycoprot... 33 0.94
SPOT_MYCPN (P75386) Probable guanosine-3',5'-bis(diphosphate) 3'... 32 3.6
Y449_MYCGE (P47687) Hypothetical protein MG449 31 4.6
MS51_YEAST (P32335) MSS51 protein 31 4.6
YA88_AQUAE (O67178) Hypothetical protein AQ_1088 31 6.1
NKGE_PANTR (Q95MI4) NKG2-E type II integral membrane protein (NK... 31 6.1
NKGE_HUMAN (Q07444) NKG2-E type II integral membrane protein (NK... 31 6.1
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 228 bits (580), Expect = 3e-59
Identities = 136/376 (36%), Positives = 210/376 (55%), Gaps = 13/376 (3%)
Query: 1 MKIPQGLEGI-PPHKVCKLDKSLYGLKQASRKWYEKLSHFLISAGYASVTSDPTLFIKR- 58
M+ P+G E H VCKL+KSLYGLKQA R+WY K F+ S Y SDP ++ KR
Sbjct: 938 MEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQTYLKTYSDPCVYFKRF 997
Query: 59 TTSTITVLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVA--HSAKG 116
+ + +LL+YVDD+++ G D I +K L F +KDLG + LG+++ +++
Sbjct: 998 SENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQQILGMKIVRERTSRK 1057
Query: 117 ITICQQQYCLDLLNKTGTLGCKPSSIPMDPSQRLHRDDSEPYVDAS------EYRALVGK 170
+ + Q++Y +L + KP S P+ +L + V+ Y + VG
Sbjct: 1058 LWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEEKGNMAKVPYSSAVGS 1117
Query: 171 LLY-LTSTRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFFPRDSSLHLL 229
L+Y + TRPDI+ V + +FL++P H++A +LRYL+ + G L F + L
Sbjct: 1118 LMYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTTGDCLCFGGSDPI-LK 1176
Query: 230 GYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALAAATCELQW 289
GY+DAD G D R+S TGY F +SW+SK Q+ V+ S++EAEY A E+ W
Sbjct: 1177 GYTDADMAGDIDNRKSSTGYLFTFSGGAISWQSKLQKCVALSTTEAEYIAATETGKEMIW 1236
Query: 290 LVYLLTDLNVTCSKVPALVCDSQSALHIASNPVFHERTKHLDIDCHVVREKLQSGLMELF 349
L L +L + K + CDSQSA+ ++ N ++H RTKH+D+ H +RE + +++
Sbjct: 1237 LKRFLQELGLH-QKEYVVYCDSQSAIDLSKNSMYHARTKHIDVRYHWIREMVDDESLKVL 1295
Query: 350 PISSQEQIADILTKAL 365
IS+ E AD+LTK +
Sbjct: 1296 KISTNENPADMLTKVV 1311
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 213 bits (542), Expect = 7e-55
Identities = 131/386 (33%), Positives = 208/386 (52%), Gaps = 8/386 (2%)
Query: 1 MKIPQGLEGIPPHKVCKLDKSLYGLKQASRKWYEKLSHFLISAGYASVTSDPTLFI--KR 58
M++PQG+ VCKL+K++YGLKQA+R W+E L + + + D ++I K
Sbjct: 1018 MRLPQGIS-CNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKG 1076
Query: 59 TTSTITVLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGIT 118
+ +L+YVDD+V+A D+ + K L F + DL +K F+G+ + I
Sbjct: 1077 NINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIY 1136
Query: 119 ICQQQYCLDLLNKTGTLGCKPSSIPMDPSQRLHRDDSEPYVDASEYRALVGKLLYLT-ST 177
+ Q Y +L+K C S P+ +S+ + + R+L+G L+Y+ T
Sbjct: 1137 LSQSAYVKKILSKFNMENCNAVSTPLPSKINYELLNSDEDCN-TPCRSLIGCLMYIMLCT 1195
Query: 178 RPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFFPRDSSLH--LLGYSDAD 235
RPD++ V L ++ + ++ +VLRYLK + L F ++ + ++GY D+D
Sbjct: 1196 RPDLTTAVNILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSD 1255
Query: 236 WGGCPDTRRSITGYCF-FIGKSLVSWKSKKQQTVSRSSSEAEYRALAAATCELQWLVYLL 294
W G R+S TGY F +L+ W +K+Q +V+ SS+EAEY AL A E WL +LL
Sbjct: 1256 WAGSEIDRKSTTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFLL 1315
Query: 295 TDLNVTCSKVPALVCDSQSALHIASNPVFHERTKHLDIDCHVVREKLQSGLMELFPISSQ 354
T +N+ + D+Q + IA+NP H+R KH+DI H RE++Q+ ++ L I ++
Sbjct: 1316 TSINIKLENPIKIYEDNQGCISIANNPSCHKRAKHIDIKYHFAREQVQNNVICLEYIPTE 1375
Query: 355 EQIADILTKALHPASFNHLIAKLSLL 380
Q+ADI TK L A F L KL LL
Sbjct: 1376 NQLADIFTKPLPAARFVELRDKLGLL 1401
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 196 bits (497), Expect = 1e-49
Identities = 96/224 (42%), Positives = 141/224 (62%), Gaps = 1/224 (0%)
Query: 66 LLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQYC 125
LL+YVDDI+L G+ + ++ L + F +KDLG + +FLG+++ G+ + Q +Y
Sbjct: 3 LLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYA 62
Query: 126 LDLLNKTGTLGCKPSSIPMDPSQRLHRDDSEPYVDASEYRALVGKLLYLTSTRPDISFTV 185
+LN G L CKP S P+ P + + Y D S++R++VG L YLT TRPDIS+ V
Sbjct: 63 EQILNNAGMLDCKPMSTPL-PLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAV 121
Query: 186 QQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRS 245
+CQ + PT F +VLRY+K + GL+ ++S L++ + D+DW GC TRRS
Sbjct: 122 NIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRS 181
Query: 246 ITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALAAATCELQW 289
TG+C F+G +++SW +K+Q TVSRSS+E EYRALA EL W
Sbjct: 182 TTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 135 bits (339), Expect = 2e-31
Identities = 90/279 (32%), Positives = 140/279 (49%), Gaps = 3/279 (1%)
Query: 12 PHKVCKLDKSLYGLKQASRKWYEKLSHFLISAGYASVTSDPTLFIKRTTSTITVLLVYVD 71
P V +L +YGLKQA W E +++ L G+ + L+ + T+ + VYVD
Sbjct: 30 PDYVWELYGGMYGLKQAPLLWNEHINNTLKKIGFCRHEGEHGLYFRSTSDGPIYIGVYVD 89
Query: 72 DIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKG-ITICQQQYCLDLLN 130
D+++A +K L ++ +KDLG + FLGL + S G IT+ Q Y +
Sbjct: 90 DLLVAAPSPKIYDRVKQELTKLYSMKDLGKVDKFLGLNIHQSTNGDITLSLQDYIAKAAS 149
Query: 131 KTGTLGCKPSSIPMDPSQRLHRDDSEPYVDASEYRALVGKLLYLTST-RPDISFTVQQLC 189
++ K + P+ S+ L S D + Y+++VG+LL+ +T RPDIS+ V L
Sbjct: 150 ESEINTFKLTQTPLCNSKPLFETTSPHLKDITPYQSIVGQLLFCANTGRPDISYPVSLLS 209
Query: 190 QFLDSPTATHFKAAHKVLRYLKTSPGTGLFFPRDSSLHLLGYSDADWGGCPDTRRSITGY 249
+FL P A H ++A +VLRYL T+ L + S + L Y DA G D S GY
Sbjct: 210 RFLREPRAIHLESARRVLRYLYTTRSMCLKYRSGSQVALTVYCDASHGAIHDLPHSTGGY 269
Query: 250 CFFIGKSLVSWKSKK-QQTVSRSSSEAEYRALAAATCEL 287
+ + V+W SKK + + S+EAEY + E+
Sbjct: 270 VTLLAGAPVTWSSKKLKGVIPVPSTEAEYITASETVMEI 308
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 105 bits (263), Expect = 1e-22
Identities = 95/383 (24%), Positives = 176/383 (45%), Gaps = 23/383 (6%)
Query: 15 VCKLDKSLYGLKQASRKWYEKLSHFLISAGYASVTSDPTLFIKRTTSTITVLLVYVDDIV 74
V KL+K+LYGLKQ+ ++W + L +L G + P L+ +T ++ VYVDD V
Sbjct: 1409 VVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY--QTEDKNLMIAVYVDDCV 1466
Query: 75 LAGNDLDEIQLIKNSLHNVFGIKDLGIL------KFFLGLEVAHSAK----GITICQQQY 124
+A ++ + N L + F +K G L LG+++ ++ + +T+
Sbjct: 1467 IAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLKSFIN 1526
Query: 125 CLDLLNKTGTLGCKPSSIP------MDPSQR-LHRDDSEPYVDASEYRALVGKLLYLT-S 176
+D + SSIP +DP + L + E + + L+G+L Y+
Sbjct: 1527 RMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFRQGVLKLQQLLGELNYVRHK 1586
Query: 177 TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFFPRDSSLH--LLGYSDA 234
R DI F V+++ + ++ P F +K+++YL G+ + RD + ++ +DA
Sbjct: 1587 CRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIAITDA 1646
Query: 235 DWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALAAATCELQWLVYLL 294
G D + I G + G ++ + S K SS+EAE A+ + + L L
Sbjct: 1647 SVGSEYDAQSRI-GVILWYGMNIFNVYSNKSTNRCVSSTEAELHAIYEGYADSETLKVTL 1705
Query: 295 TDLNVTCSKVPALVCDSQSALHIASNPVFHERTKHLDIDCHVVREKLQSGLMELFPISSQ 354
+L + ++ DS+ A+ + + K I +++EK++ ++L I+ +
Sbjct: 1706 KELGEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLKITGK 1765
Query: 355 EQIADILTKALHPASFNHLIAKL 377
IAD+LTK + + F I L
Sbjct: 1766 GNIADLLTKPVSASDFKRFIQVL 1788
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 97.1 bits (240), Expect = 7e-20
Identities = 100/396 (25%), Positives = 180/396 (45%), Gaps = 38/396 (9%)
Query: 11 PPH-----KVCKLDKSLYGLKQASRKWYEKLSHFLI-SAGYASVTSDPTLFIKRTTSTIT 64
PPH K+ +L KSLYGLKQ+ WYE + +LI G V +F S +T
Sbjct: 938 PPHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF---KNSQVT 994
Query: 65 VLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQY 124
+ L +VDD++L DL+ + I +L + K + + + E+ + G+ I Q+
Sbjct: 995 ICL-FVDDMILFSKDLNANKKIITTLKKQYDTKIINLGE--SDNEIQYDILGLEIKYQRG 1051
Query: 125 CLDLLNKTGTLGCK--PSSIPMDPSQR---------LHRDDSEPYVDASEYRALV---GK 170
L +L K ++P++P R L+ D E +D EY+ V K
Sbjct: 1052 KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQK 1111
Query: 171 LLYLTS-----TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFF----P 221
L+ L S R D+ + + L Q + P+ +++++++ + L + P
Sbjct: 1112 LIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKP 1171
Query: 222 RDSSLHLLGYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALA 281
+ L+ SDA +G P + I G + + ++ KS K S++EAE A++
Sbjct: 1172 TEPDNKLVAISDASYGNQPYYKSQI-GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAIS 1230
Query: 282 AATCELQWLVYLLTDLNVTCSKVPALVCDSQSALHIASNPVFHE-RTKHLDIDCHVVREK 340
+ L L YL+ +LN + L+ DS+S + I + + R + +R++
Sbjct: 1231 ESVPLLNNLSYLIQELNKK-PIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDE 1289
Query: 341 LQSGLMELFPISSQEQIADILTKALHPASFNHLIAK 376
+ + ++ I +++ IAD++TK L +F L K
Sbjct: 1290 VSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 95.5 bits (236), Expect = 2e-19
Identities = 100/396 (25%), Positives = 179/396 (44%), Gaps = 38/396 (9%)
Query: 11 PPH-----KVCKLDKSLYGLKQASRKWYEKLSHFLI-SAGYASVTSDPTLFIKRTTSTIT 64
PPH K+ +L KS YGLKQ+ WYE + +LI G V +F S +T
Sbjct: 1365 PPHLGMNDKLIRLKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF---KNSQVT 1421
Query: 65 VLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQY 124
+ L +VDD++L DL+ + I +L + K + + + E+ + G+ I Q+
Sbjct: 1422 ICL-FVDDMILFSKDLNANKKIITTLKKQYDTKIINLGE--SDNEIQYDILGLEIKYQRG 1478
Query: 125 CLDLLNKTGTLGCK--PSSIPMDPSQR---------LHRDDSEPYVDASEYRALV---GK 170
L +L K ++P++P R L+ D E +D EY+ V K
Sbjct: 1479 KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQK 1538
Query: 171 LLYLTS-----TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFF----P 221
L+ L S R D+ + + L Q + P+ +++++++ + L + P
Sbjct: 1539 LIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKP 1598
Query: 222 RDSSLHLLGYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALA 281
+ L+ SDA +G P + I G F + ++ KS K S++EAE A++
Sbjct: 1599 TEPDNKLVAISDASYGNQPYYKSQI-GNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAIS 1657
Query: 282 AATCELQWLVYLLTDLNVTCSKVPALVCDSQSALHIASNPVFHE-RTKHLDIDCHVVREK 340
+ L L YL+ +LN + L+ DS+S + I + + R + +R++
Sbjct: 1658 ESVPLLNNLSYLIQELNKK-PIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDE 1716
Query: 341 LQSGLMELFPISSQEQIADILTKALHPASFNHLIAK 376
+ + ++ I +++ IAD++TK L +F L K
Sbjct: 1717 VSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1752
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 94.4 bits (233), Expect = 4e-19
Identities = 103/399 (25%), Positives = 180/399 (44%), Gaps = 44/399 (11%)
Query: 11 PPH-----KVCKLDKSLYGLKQASRKWYEKLSHFLI-SAGYASVTSDPTLFIKRTTSTIT 64
PPH K+ +L KSLYGLKQ+ WYE + +LI G V +F S +T
Sbjct: 938 PPHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF---KNSQVT 994
Query: 65 VLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQY 124
+ L +VDD++L DL+ + I +L + K + + + E+ + G+ I Q+
Sbjct: 995 ICL-FVDDMILFSKDLNANKKIITTLKKQYDTKIINLGE--SDNEIQYDILGLEIKYQRG 1051
Query: 125 CLDLLNKTGTLGCK--PSSIPMDPSQR---------LHRDDSEPYVDASEYRALV---GK 170
L +L K ++P++P R L+ D E +D EY+ V K
Sbjct: 1052 KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYKEKVHEMQK 1111
Query: 171 LLYLTS-----TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFF----P 221
L+ L S R D+ + + L Q + P+ +++++++ + L + P
Sbjct: 1112 LIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKP 1171
Query: 222 RDSSLHLLGYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALA 281
+ L+ SDA +G P + I G + + ++ KS K S++EAE A++
Sbjct: 1172 TEPDNKLVAISDASYGNQPYYKSQI-GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAIS 1230
Query: 282 AATCELQWLVYLLTDLNVTCSKVP---ALVCDSQSALH-IASNPVFHERTKHLDIDCHVV 337
+ L L +L+ +LN K P L+ DS+S + I SN R + +
Sbjct: 1231 ESVPLLNNLSHLVQELN----KKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRL 1286
Query: 338 REKLQSGLMELFPISSQEQIADILTKALHPASFNHLIAK 376
R+++ + + I +++ IAD++TK L +F L K
Sbjct: 1287 RDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 93.6 bits (231), Expect = 8e-19
Identities = 99/400 (24%), Positives = 176/400 (43%), Gaps = 46/400 (11%)
Query: 11 PPH-----KVCKLDKSLYGLKQASRKWYEKLSHFLISAGYASVTSDPTLFIKRTTSTITV 65
PPH K+ +L KSLYGLKQ+ WYE + +LI+ + K + TI
Sbjct: 1380 PPHLGLNDKLLRLRKSLYGLKQSGANWYETIKSYLINCCDMQEVRGWSCVFKNSQVTI-- 1437
Query: 66 LLVYVDDIVLAGNDLDEIQLIKNSLHNVF-------GIKDLGILKFFLGLEVAHSAKGIT 118
++VDD++L DL+ + I +L + G D I LGLE+ +
Sbjct: 1438 -CLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQ----- 1491
Query: 119 ICQQQYCLDLLNKTGTLGCKPSSIPMDPSQRLHR---------DDSEPYVDASEYRALV- 168
+ +Y + K+ T ++P++P + R D E +D EY+ V
Sbjct: 1492 --RSKYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHYIDQDELEIDEDEYKEKVH 1549
Query: 169 --GKLLYLTS-----TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFF- 220
KL+ L S R D+ + + L Q + P+ +++++++ + L +
Sbjct: 1550 EMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWH 1609
Query: 221 ---PRDSSLHLLGYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEY 277
P L+ SDA +G P + I G F + ++ KS K S++EAE
Sbjct: 1610 KNKPTKPDNKLVAISDASYGNQPYYKSQI-GNIFLLNGKVIGGKSTKASLTCTSTTEAEI 1668
Query: 278 RALAAATCELQWLVYLLTDLNVTCSKVPALVCDSQSALHIASNPVFHE-RTKHLDIDCHV 336
A++ A L L +L+ +LN + L+ DS+S + I + + R +
Sbjct: 1669 HAVSEAIPLLNNLSHLVQELNKK-PIIKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMR 1727
Query: 337 VREKLQSGLMELFPISSQEQIADILTKALHPASFNHLIAK 376
+R+++ + ++ I +++ IAD++TK L +F L K
Sbjct: 1728 LRDEVSGNNLYVYYIETKKNIADVMTKPLPIKTFKLLTNK 1767
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 89.7 bits (221), Expect = 1e-17
Identities = 38/82 (46%), Positives = 54/82 (65%)
Query: 172 LYLTSTRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFFPRDSSLHLLGY 231
+YLT TRPD++F V +L QF + +A +KVL Y+K + G GLF+ S L L +
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 232 SDADWGGCPDTRRSITGYCFFI 253
+D+DW CPDTRRS+TG+C +
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLV 82
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 88.6 bits (218), Expect = 2e-17
Identities = 101/399 (25%), Positives = 180/399 (44%), Gaps = 44/399 (11%)
Query: 11 PPH-----KVCKLDKSLYGLKQASRKWYEKLSHFLI-SAGYASVTSDPTLFIKRTTSTIT 64
PPH K+ +L KSLYGLKQ+ WYE + +LI G V +F S +T
Sbjct: 938 PPHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF---KNSQVT 994
Query: 65 VLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQY 124
+ L +VDD++L DL+ + I +L + K + + + E+ + G+ I Q+
Sbjct: 995 ICL-FVDDMILFSKDLNANKKIITTLKKQYDTKIINLGE--SDNEIQYDILGLEIKYQRG 1051
Query: 125 CLDLLNKTGTLGCK--PSSIPMDPSQR---------LHRDDSEPYVDASEYRALV---GK 170
L +L K ++P++P R L+ D E ++ +Y+ V K
Sbjct: 1052 KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQK 1111
Query: 171 LLYLTS-----TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFF----P 221
L+ L S R D+ + + L Q + P+ +++++++ + L + P
Sbjct: 1112 LIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKP 1171
Query: 222 RDSSLHLLGYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALA 281
+ L+ SDA +G P + I G + + ++ KS K S++EAE A++
Sbjct: 1172 VKPTNKLVVISDASYGNQPYYKSQI-GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAIS 1230
Query: 282 AATCELQWLVYLLTDLNVTCSKVP---ALVCDSQSALH-IASNPVFHERTKHLDIDCHVV 337
+ L L +L+ +LN K P L+ DS+S + I SN R + +
Sbjct: 1231 ESVPLLNNLSHLVQELN----KKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRL 1286
Query: 338 REKLQSGLMELFPISSQEQIADILTKALHPASFNHLIAK 376
R+++ + + I +++ IAD++TK L +F L K
Sbjct: 1287 RDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 87.0 bits (214), Expect = 7e-17
Identities = 101/399 (25%), Positives = 180/399 (44%), Gaps = 44/399 (11%)
Query: 11 PPH-----KVCKLDKSLYGLKQASRKWYEKLSHFLI-SAGYASVTSDPTLFIKRTTSTIT 64
PPH K+ +L KSLYGLKQ+ WYE + +LI G V +F S +T
Sbjct: 938 PPHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF---ENSQVT 994
Query: 65 VLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQY 124
+ L +VDD+VL +L+ + I + L + K + + + E+ + G+ I Q+
Sbjct: 995 ICL-FVDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGE--SDEEIQYDILGLEIKYQRG 1051
Query: 125 CLDLLNKTGTLGCK--PSSIPMDPSQR---------LHRDDSEPYVDASEYRALV---GK 170
L +L K ++P++P R L+ D E ++ +Y+ V K
Sbjct: 1052 KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQK 1111
Query: 171 LLYLTS-----TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFF----P 221
L+ L S R D+ + + L Q + P+ +++++++ + L + P
Sbjct: 1112 LIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKP 1171
Query: 222 RDSSLHLLGYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALA 281
+ L+ SDA +G P + I G + + ++ KS K S++EAE A++
Sbjct: 1172 VKPTNKLVVISDASYGNQPYYKSQI-GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAIS 1230
Query: 282 AATCELQWLVYLLTDLNVTCSKVP---ALVCDSQSALH-IASNPVFHERTKHLDIDCHVV 337
+ L L YL+ +L+ K P L+ DS+S + I SN R + +
Sbjct: 1231 ESVPLLNNLSYLIQELD----KKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRL 1286
Query: 338 REKLQSGLMELFPISSQEQIADILTKALHPASFNHLIAK 376
R+++ + + I +++ IAD++TK L +F L K
Sbjct: 1287 RDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1325
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 86.7 bits (213), Expect = 9e-17
Identities = 101/399 (25%), Positives = 179/399 (44%), Gaps = 44/399 (11%)
Query: 11 PPH-----KVCKLDKSLYGLKQASRKWYEKLSHFLI-SAGYASVTSDPTLFIKRTTSTIT 64
PPH K+ +L KSLYGLKQ+ WYE + +LI G V +F S +T
Sbjct: 1365 PPHLGMNDKLIRLKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWSCVF---KNSQVT 1421
Query: 65 VLLVYVDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQY 124
+ L +VDD+VL +L+ + I L + K + + + E+ + G+ I Q+
Sbjct: 1422 ICL-FVDDMVLFSKNLNSNKRIIEKLKMQYDTKIINLGE--SDEEIQYDILGLEIKYQRG 1478
Query: 125 CLDLLNKTGTLGCK--PSSIPMDPSQR---------LHRDDSEPYVDASEYRALV---GK 170
L +L K ++P++P R L+ D E ++ +Y+ V K
Sbjct: 1479 KYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQK 1538
Query: 171 LLYLTS-----TRPDISFTVQQLCQFLDSPTATHFKAAHKVLRYLKTSPGTGLFF----P 221
L+ L S R D+ + + L Q + P+ +++++++ + L + P
Sbjct: 1539 LIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKP 1598
Query: 222 RDSSLHLLGYSDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEAEYRALA 281
+ L+ SDA +G P + I G + + ++ KS K S++EAE A++
Sbjct: 1599 VKPTNKLVVISDASYGNQPYYKSQI-GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAIS 1657
Query: 282 AATCELQWLVYLLTDLNVTCSKVP---ALVCDSQSALH-IASNPVFHERTKHLDIDCHVV 337
+ L L YL+ +L+ K P L+ DS+S + I SN R + +
Sbjct: 1658 ESVPLLNNLSYLIQELD----KKPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRL 1713
Query: 338 REKLQSGLMELFPISSQEQIADILTKALHPASFNHLIAK 376
R+++ + + I +++ IAD++TK L +F L K
Sbjct: 1714 RDEVSGNHLHVCYIETKKNIADVMTKPLPIKTFKLLTNK 1752
>VGLM_BUNGE (P12430) M polyprotein precursor [Contains: Glycoprotein
G2; Nonstructural protein NS-M; Glycoprotein G1]
Length = 1437
Score = 33.5 bits (75), Expect = 0.94
Identities = 26/85 (30%), Positives = 42/85 (48%), Gaps = 13/85 (15%)
Query: 70 VDDIVLAGNDLDEIQLIKNSLHNVFGIKDLGILKFFLGLEVAHSAKGITICQQQYCLDLL 129
V+DIVL G D++ Q +K+S+ + I +GI+ F GL V + L+L+
Sbjct: 351 VNDIVL-GRDME--QELKSSILILMSICGIGIILIFFGLTVL----------LEIVLELI 397
Query: 130 NKTGTLGCKPSSIPMDPSQRLHRDD 154
K T+ CK ++ D +R D
Sbjct: 398 AKRSTIFCKECNLIHDKKSMTYRGD 422
>SPOT_MYCPN (P75386) Probable guanosine-3',5'-bis(diphosphate)
3'-pyrophosphohydrolase (EC 3.1.7.2) ((ppGpp)ase)
(Penta-phosphate guanosine-3'-pyrophosphohydrolase)
Length = 733
Score = 31.6 bits (70), Expect = 3.6
Identities = 26/130 (20%), Positives = 61/130 (46%), Gaps = 22/130 (16%)
Query: 258 VSWKSKKQQTVSRSSSEAEYRALA----AATCELQWLVYLLTDLNVTCSKVPALVCDSQS 313
++ +SKKQ+ ++R + ++L ++ E+ LV L D S + L + Q
Sbjct: 120 ITSESKKQRQLNRKKEDLNLKSLVNIAMSSQQEVNALVLKLADRLDNISSIEFLAVEKQK 179
Query: 314 ALHIASNPVFHERTKHLDIDCHVVREKLQSGLMELFPISSQEQIADILTKALHPASFNHL 373
+ + L++ + +G + ++P+ +Q +AD+ K L P +FN+
Sbjct: 180 II----------AKETLELYAKI------AGRIGMYPVKTQ--LADLSFKVLDPKNFNNT 221
Query: 374 IAKLSLLNIY 383
++K++ ++
Sbjct: 222 LSKINQQKVF 231
>Y449_MYCGE (P47687) Hypothetical protein MG449
Length = 228
Score = 31.2 bits (69), Expect = 4.6
Identities = 35/109 (32%), Positives = 48/109 (43%), Gaps = 14/109 (12%)
Query: 232 SDADWGGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSSEA-EYRALAAATCELQWL 290
+D DW + ITG+ FF K KS K+ +S +E Y +L + L
Sbjct: 38 NDKDWSFFVNDANRITGFNFFNIK-----KSFKKFFLSHRFNEGLNYPSLKIMKRISELL 92
Query: 291 VYLLTDLNVTCSKVPALVCDSQSALHIASN-----PVFHERTKHLDIDC 334
Y DL +KVP +VC+ S + IA+ V TK LDI C
Sbjct: 93 GY---DLISLANKVPFVVCEVVSVIPIANTHLKRCKVNTGLTKSLDIVC 138
>MS51_YEAST (P32335) MSS51 protein
Length = 436
Score = 31.2 bits (69), Expect = 4.6
Identities = 19/52 (36%), Positives = 24/52 (45%), Gaps = 2/52 (3%)
Query: 135 LGCKPSSIPMDPS--QRLHRDDSEPYVDASEYRALVGKLLYLTSTRPDISFT 184
LG P P DP+ R H D P VD E A + L + T DI++T
Sbjct: 44 LGLDPPPSPEDPTPENRFHPWDQSPSVDLRERAAKIRTLAHCPVTGKDINYT 95
>YA88_AQUAE (O67178) Hypothetical protein AQ_1088
Length = 761
Score = 30.8 bits (68), Expect = 6.1
Identities = 13/24 (54%), Positives = 19/24 (79%)
Query: 84 QLIKNSLHNVFGIKDLGILKFFLG 107
Q+ +NSL++ G+ DLG+L FFLG
Sbjct: 60 QVSENSLNSAQGLSDLGLLYFFLG 83
>NKGE_PANTR (Q95MI4) NKG2-E type II integral membrane protein
(NKG2-E activating NK receptor) (NK cell receptor E)
Length = 240
Score = 30.8 bits (68), Expect = 6.1
Identities = 12/37 (32%), Positives = 21/37 (56%)
Query: 237 GGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSS 273
G CP+ + + C++IGK +W+ Q S++SS
Sbjct: 115 GHCPEEWITYSNSCYYIGKERRTWEESLQACASKNSS 151
>NKGE_HUMAN (Q07444) NKG2-E type II integral membrane protein
(NKG2-E activating NK receptor) (NK cell receptor E)
Length = 240
Score = 30.8 bits (68), Expect = 6.1
Identities = 12/37 (32%), Positives = 21/37 (56%)
Query: 237 GGCPDTRRSITGYCFFIGKSLVSWKSKKQQTVSRSSS 273
G CP+ + + C++IGK +W+ Q S++SS
Sbjct: 115 GHCPEEWITYSNSCYYIGKERRTWEESLQACASKNSS 151
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.136 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 44,633,955
Number of Sequences: 164201
Number of extensions: 1784353
Number of successful extensions: 3993
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 3942
Number of HSP's gapped (non-prelim): 24
length of query: 385
length of database: 59,974,054
effective HSP length: 112
effective length of query: 273
effective length of database: 41,583,542
effective search space: 11352306966
effective search space used: 11352306966
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Medicago: description of AC146910.6


