

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.5 - phase: 0 /pseudo
(470 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
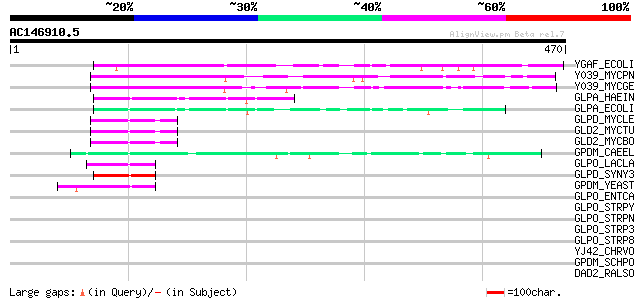
Score E
Sequences producing significant alignments: (bits) Value
YGAF_ECOLI (P37339) Hypothetical protein ygaF 108 2e-23
Y039_MYCPN (P75063) Hypothetical protein MG039 homolog (D09_orf384) 81 7e-15
Y039_MYCGE (P47285) Hypothetical protein MG039 76 2e-13
GLPA_HAEIN (P43799) Anaerobic glycerol-3-phosphate dehydrogenase... 52 3e-06
GLPA_ECOLI (P13032) Anaerobic glycerol-3-phosphate dehydrogenase... 52 4e-06
GLPD_MYCLE (P53435) Glycerol-3-phosphate dehydrogenase (EC 1.1.9... 47 8e-05
GLD2_MYCTU (P64184) Glycerol-3-phosphate dehydrogenase 2 (EC 1.1... 47 8e-05
GLD2_MYCBO (P64185) Glycerol-3-phosphate dehydrogenase 2 (EC 1.1... 47 8e-05
GPDM_CAEEL (P90795) Probable glycerol-3-phosphate dehydrogenase,... 47 1e-04
GLPO_LACLA (Q9CG65) Alpha-glycerophosphate oxidase (EC 1.1.3.21)... 46 2e-04
GLPD_SYNY3 (P74257) Glycerol-3-phosphate dehydrogenase (EC 1.1.9... 45 5e-04
GPDM_YEAST (P32191) Glycerol-3-phosphate dehydrogenase, mitochon... 44 7e-04
GLPO_ENTCA (O86963) Alpha-glycerophosphate oxidase (EC 1.1.3.21)... 44 0.001
GLPO_STRPY (Q99YI8) Alpha-glycerophosphate oxidase (EC 1.1.3.21)... 43 0.002
GLPO_STRPN (P35596) Alpha-glycerophosphate oxidase (EC 1.1.3.21)... 43 0.002
GLPO_STRP3 (Q8K666) Alpha-glycerophosphate oxidase (EC 1.1.3.21)... 43 0.002
GLPO_STRP8 (Q8NZX0) Alpha-glycerophosphate oxidase (EC 1.1.3.21)... 43 0.002
YJ42_CHRVO (Q7NWN9) Hypothetical UPF0209 protein CV1942 42 0.003
GPDM_SCHPO (O14400) Glycerol-3-phosphate dehydrogenase, mitochon... 42 0.003
DAD2_RALSO (Q8XX54) D-amino acid dehydrogenase 2 small subunit (... 42 0.003
>YGAF_ECOLI (P37339) Hypothetical protein ygaF
Length = 444
Score = 108 bits (271), Expect = 2e-23
Identities = 105/426 (24%), Positives = 178/426 (41%), Gaps = 64/426 (15%)
Query: 72 DCVVIGAGVVGIAVARAL--ALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSLK 129
D V+IG G++G++ A L + ++E + + NS V+HAG+YY SLK
Sbjct: 25 DFVIIGGGIIGMSTAMQLIDVYPDARIALLEKESAPACHQTGHNSGVIHAGVYYTPGSLK 84
Query: 130 AIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMDG 189
A FC+ G +C ++ I ++ GK++VAT E+ ++ + NG++ + ++
Sbjct: 85 AQFCLAGNRATKAFCDQNGIRYDNCGKMLVATSDLEMERMRALWERTAANGIE-REWLNA 143
Query: 190 VDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTYN 249
+ + EP + + I P SGIV + ++ ++ G YN
Sbjct: 144 DELREREPNITGLGGIFVPSSGIVSYRDVTAAMAKIF-------------QSRGGEIIYN 190
Query: 250 STVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFTG 309
+ V G + N + V T+ E+ + +++ +GL A L K G
Sbjct: 191 AEVSGLNEHKNGV---VIRTRQGGEYEAST------------LISCSGLMADRLVK-MLG 234
Query: 310 LENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDG--GLGVHVTLDLNGQVKFGP-- 365
LE I + RG YF L+ HLIYPIP+ LGVH+T ++G V GP
Sbjct: 235 LEPGFIICPF--RGEYFRLAPEHNQIVNHLIYPIPDPAMPFLGVHLTRMIDGSVTVGPNA 292
Query: 366 ------------DVEWIDGVDDISS------FQNKFDYSVQANR----AEKFYPEIRKYY 403
D + D ++ + S QN + + + ++KY
Sbjct: 293 VLAFKREGYRKRDFSFSDTLEILGSSGIRRVLQNHLRSGLGEMKNSLCKSGYLRLVQKYC 352
Query: 404 PNLKDGSLEPGYSGIRPKLSGPCQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
P L L+P +G+R + P +D + P I+ SP TS++ I
Sbjct: 353 PRLSLSDLQPWPAGVRAQAVSPDGKLIDDFL----FVTTPRTIHTCNAPSPAATSAIPIG 408
Query: 464 DFISTK 469
I +K
Sbjct: 409 AHIVSK 414
>Y039_MYCPN (P75063) Hypothetical protein MG039 homolog (D09_orf384)
Length = 384
Score = 80.9 bits (198), Expect = 7e-15
Identities = 96/406 (23%), Positives = 166/406 (40%), Gaps = 62/406 (15%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E D +++G GV+G A A L+ +V ++E TS NS V+H GI H L
Sbjct: 2 ETRDVLIVGGGVIGCATAYELSQYKLKVTLVEKHHYLAQETSHANSGVIHTGIDPNPHKL 61
Query: 129 KAIFCVKGREM-LYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGV--DGLK 185
A + + G+++ L Y + P ++ LIVA E +L V+ GI N + + ++
Sbjct: 62 TAKYNILGKKLWLNTYFKRLGFPRQKIRTLIVAFNEMEREQLEVLKQRGIANQINLEDIQ 121
Query: 186 MMDGVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGAT 245
M+ + +K+EP + + ++ L + I+P+L
Sbjct: 122 MLSKEETLKLEPYV---------------NPEIVAGLKIEGSWAIDPVLAS--------- 157
Query: 246 FTYNSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIP----KLVVNSAG---- 297
+ +IC + T K+ +G + P K ++++AG
Sbjct: 158 ---KCLALAAQQNKVQICTNTEVTNISKQVDGTYLVWTNNETTPSFKVKKIIDAAGHYAD 214
Query: 298 -LSALALAKRFTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLD 356
L+ LA A F RG Y ++N +++ +P G GV V+
Sbjct: 215 YLAHLAKADDFEQTTR---------RGQYVVVTNQGELHLNSMVFMVPTIHGKGVIVSPM 265
Query: 357 LNGQVKFGPDVEWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYS 416
L+G GP +DGVD ++ D + K P+L + ++
Sbjct: 266 LDGNFLVGPTA--LDGVDKEATRYITKDAPCMLTKIGK------HMVPSLNINNALISFA 317
Query: 417 GIRPKLSGPCQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAI 462
G RP ++ DF+I+ H P + L G++SPGLT++ AI
Sbjct: 318 GSRP----IDKATNDFIIR--VAHNDPDFVILGGMKSPGLTAAPAI 357
>Y039_MYCGE (P47285) Hypothetical protein MG039
Length = 384
Score = 76.3 bits (186), Expect = 2e-13
Identities = 97/403 (24%), Positives = 184/403 (45%), Gaps = 54/403 (13%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
+ +D +++G GV+G + A L+ +V ++E G TS NS V+H+GI + L
Sbjct: 2 QTIDVLIVGGGVIGTSCAYELSQYKLKVALLEKNAFLGCETSQANSGVIHSGIDPNPNKL 61
Query: 129 KAIFCVKGREM-LYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNG--VDGLK 185
A + + GR++ + ++ K P ++ LIVA + E +L+++ GI+N V+ ++
Sbjct: 62 TAKYNILGRKIWIEDWFKKLIFPRKKIATLIVAFNNEEKLQLNLLKERGIKNSIPVENIQ 121
Query: 186 MMDGVDAMKMEPELQCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPM-----LVQGEAE 240
++D + EP ++P +++ SL + I+P+ L +
Sbjct: 122 ILDQQQTLLQEP-------FINP--------NVVASLKVEGSWLIDPLIATKCLALASLQ 166
Query: 241 NHGATFTYNSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSA 300
N+ A ++ N V ++ ++ L ++ ++ K ++++AG A
Sbjct: 167 NNVAIYS-NKKVTKIEIDSDDDFLVFINNETTPQFKTKK------------LIDAAGHYA 213
Query: 301 LALAKRFTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHVTLDLNGQ 360
LA+ T ++N +G Y L N +I+ +P G GV V L+G
Sbjct: 214 DWLAET-TQVDNF---KQTTRKGQYLVLKNQNNLKINTIIFMVPTIHGKGVVVAEMLDGN 269
Query: 361 VKFGPDVEWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRP 420
+ GP+ ++G++ +NK S+ + + +K P+L+ + ++G R
Sbjct: 270 ILVGPNA--VEGIE-----KNK-TRSIDLDSINQIKTIGKKMVPSLQFENSIYSFAGSR- 320
Query: 421 KLSGPCQSPVDFVIQGEDIHGVPGLINLFGIESPGLTSSLAIA 463
DFVI+ + P I L G++SPGLTSS AIA
Sbjct: 321 ---AIDIETNDFVIRTAKSN--PNFIILGGMKSPGLTSSPAIA 358
>GLPA_HAEIN (P43799) Anaerobic glycerol-3-phosphate dehydrogenase
subunit A (EC 1.1.99.5) (G-3-P dehydrogenase)
Length = 563
Score = 52.4 bits (124), Expect = 3e-06
Identities = 46/173 (26%), Positives = 79/173 (45%), Gaps = 9/173 (5%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYP-HHSLKA 130
D ++IG G G +AR AL+G + I++E TG + RN ++H+G Y + A
Sbjct: 20 DVIIIGGGATGAGIARDCALRGIDCILLERR-DIATGATGRNHGLLHSGARYAVNDQESA 78
Query: 131 IFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMDGV 190
C+K +L + A+H + ++T L + + L ++G+D + +D
Sbjct: 79 EECIKENRILRK-IARHCV--DETEGLFITLPEDSLDYQKTFLESCAKSGIDA-QAIDPE 134
Query: 191 DAMKMEPEL--QCVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAEN 241
A MEP + V A++ P G +D L S ++ N M E +N
Sbjct: 135 LAKIMEPSVNPDLVGAVVVP-DGSIDPFRLTASNMMDATENGAKMFTYCEVKN 186
>GLPA_ECOLI (P13032) Anaerobic glycerol-3-phosphate dehydrogenase
subunit A (EC 1.1.99.5) (G-3-P dehydrogenase)
Length = 542
Score = 51.6 bits (122), Expect = 4e-06
Identities = 81/355 (22%), Positives = 139/355 (38%), Gaps = 54/355 (15%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYP-HHSLKA 130
D ++IG G G +AR AL+G VI++E TG + RN ++H+G Y + A
Sbjct: 10 DVIIIGGGATGAGIARDCALRGLRVILVE-RHDIATGATGRNHGLLHSGARYAVTDAESA 68
Query: 131 IFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIPKLSVILNHGIQNGVDGLKMMDGV 190
C+ ++L + A+H + E T L + ++ + + + G+ + +D
Sbjct: 69 RECISENQIL-KRIARHCV--EPTNGLFITLPEDDLSFQATFIRACEEAGISA-EAIDPQ 124
Query: 191 DAMKMEPELQ--CVKAILSPLSGIVDSHSLMLSLVLHIDRNINPMLVQGEAENHGATFTY 248
A +EP + + A+ P G VD L + +L +A+ HGA
Sbjct: 125 QARIIEPAVNPALIGAVKVP-DGTVDPFRLTAANML-------------DAKEHGAVILT 170
Query: 249 NSTVIGGHMEGNEICLHVSETKSLKEWNGKSSLQPELVLIPKLVVNSAGLSALALAKRFT 308
V G EG +C + G++ L +VVN+AG+ +A+ +
Sbjct: 171 AHEVTGLIREGATVC----GVRVRNHLTGETQ-----ALHAPVVVNAAGIWGQHIAE-YA 220
Query: 309 GLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPIPEDGGLGVHV---TLDLNGQVKFGP 365
L ++ P A+G + + +H+I + + V T+ L G
Sbjct: 221 DLRIRMFP----AKGSLLIMDHRIN---QHVINRCRKPSDADILVPGDTISLIGTTSLRI 273
Query: 366 DVEWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYYPNLKDGSLEPGYSGIRP 420
D ID D V A + E K P + + YSG+RP
Sbjct: 274 DYNEID------------DNRVTAEEVDILLREGEKLAPVMAKTRILRAYSGVRP 316
>GLPD_MYCLE (P53435) Glycerol-3-phosphate dehydrogenase (EC
1.1.99.5)
Length = 585
Score = 47.4 bits (111), Expect = 8e-05
Identities = 31/74 (41%), Positives = 43/74 (57%), Gaps = 5/74 (6%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E+ D VVIG GVVG A A +G +V ++E A +GTSSR+S++ H G+ Y L
Sbjct: 34 EQFDVVVIGGGVVGSGCALDAATRGLKVALVE-ARDLASGTSSRSSKMFHGGLRY----L 88
Query: 129 KAIFCVKGREMLYE 142
+ + RE LYE
Sbjct: 89 EQLEFGLVREALYE 102
>GLD2_MYCTU (P64184) Glycerol-3-phosphate dehydrogenase 2 (EC
1.1.99.5)
Length = 585
Score = 47.4 bits (111), Expect = 8e-05
Identities = 31/74 (41%), Positives = 43/74 (57%), Gaps = 5/74 (6%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E+ D VVIG GVVG A A +G +V ++E A +GTSSR+S++ H G+ Y L
Sbjct: 34 EQFDVVVIGGGVVGSGCALDAATRGLKVALVE-ARDLASGTSSRSSKMFHGGLRY----L 88
Query: 129 KAIFCVKGREMLYE 142
+ + RE LYE
Sbjct: 89 EQLEFGLVREALYE 102
>GLD2_MYCBO (P64185) Glycerol-3-phosphate dehydrogenase 2 (EC
1.1.99.5)
Length = 585
Score = 47.4 bits (111), Expect = 8e-05
Identities = 31/74 (41%), Positives = 43/74 (57%), Gaps = 5/74 (6%)
Query: 69 ERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPHHSL 128
E+ D VVIG GVVG A A +G +V ++E A +GTSSR+S++ H G+ Y L
Sbjct: 34 EQFDVVVIGGGVVGSGCALDAATRGLKVALVE-ARDLASGTSSRSSKMFHGGLRY----L 88
Query: 129 KAIFCVKGREMLYE 142
+ + RE LYE
Sbjct: 89 EQLEFGLVREALYE 102
>GPDM_CAEEL (P90795) Probable glycerol-3-phosphate dehydrogenase,
mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M)
Length = 722
Score = 46.6 bits (109), Expect = 1e-04
Identities = 89/411 (21%), Positives = 160/411 (38%), Gaps = 44/411 (10%)
Query: 52 TSYPVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSS 111
++ P ++ T+ S E D ++IG G G VA +G + ++E F +GTSS
Sbjct: 57 SALPTRKDILTNLS-KGEEFDVLIIGGGATGAGVALDAQTRGLKTALVE-LDDFSSGTSS 114
Query: 112 RNSEVVHAGIYYPHHSLKAIFCVKGREMLYEYCAKHDIPHEQTGKLIVATRSSEIP-KLS 170
R+++++H G+ Y ++ + + R + +H++ I SS +P L
Sbjct: 115 RSTKLIHGGVRYLQAAIMKLDLEQYRMVKEALFERHNLLE------IAPHLSSPLPIMLP 168
Query: 171 VILNHGIQNGVDGLKMMDGVDAMK-MEPELQCVKAILSPLSGIVDSHSLMLSLVL----H 225
+ + G+K D V + ++ K+ ++ + SL +L+ H
Sbjct: 169 IYKLWQVPYYWSGIKAYDFVSGKRVLKNSFFINKSQALERFPMLRNESLKGALIYYDGQH 228
Query: 226 IDRNINPMLVQGEAENHGATFTYNSTV--IGGHMEGNEICLHVSETKSLKEWNGKSSLQP 283
D +N ++ A HGA + V + G I HV + + EW+ K+
Sbjct: 229 NDARMNLAIIL-TAIRHGAACANHVRVEKLNKDETGKVIGAHVRDMVTGGEWDIKA---- 283
Query: 284 ELVLIPKLVVNSAGLSALALAKRFTGLENKVIPPAYYARGCYFTLSNTKASPFRHLIYPI 343
K V+N+ G ++ R G P + G + TL + L+ P
Sbjct: 284 ------KAVINATGPFTDSI--RLMGDPETARPICAPSSGVHITLPGYYSPSNTGLLDPD 335
Query: 344 PEDGGLGVHVTLDLNGQVKFGPDVEWIDGVDDISSFQNKFDYSVQANRAEKFYPEIRKYY 403
DG V L G D D++ D+ + E EIR Y
Sbjct: 336 TSDG--RVIFFLPWERMTIAGT----TDAPSDVTLSPQPTDHDI-----EFILQEIRGYL 384
Query: 404 P---NLKDGSLEPGYSGIRPKLSGPCQSPVDFVIQGEDIH-GVPGLINLFG 450
+++ G + +SG+RP + P + + + I G GLI + G
Sbjct: 385 SKDVSVRRGDVMSAWSGLRPLVRDPNKKDTKSLARNHIIEVGKSGLITIAG 435
>GLPO_LACLA (Q9CG65) Alpha-glycerophosphate oxidase (EC 1.1.3.21)
(Glycerol-3-phosphate oxidase)
Length = 609
Score = 45.8 bits (107), Expect = 2e-04
Identities = 23/58 (39%), Positives = 34/58 (57%), Gaps = 1/58 (1%)
Query: 66 VPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYY 123
+ E +D +VIG G+ G + A G +V V+E F GTSSR++++VH GI Y
Sbjct: 15 IQNEEMDLLVIGGGITGAGLTLQAAAAGMKVAVLE-MQDFSEGTSSRSTKLVHGGIRY 71
>GLPD_SYNY3 (P74257) Glycerol-3-phosphate dehydrogenase (EC
1.1.99.5)
Length = 553
Score = 44.7 bits (104), Expect = 5e-04
Identities = 21/52 (40%), Positives = 34/52 (65%), Gaps = 1/52 (1%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYY 123
D +VIG G+ G+ AR AL+G + ++IE F +GTSS ++ ++H G+ Y
Sbjct: 13 DLIVIGGGINGVGTARDGALRGLKTLLIEK-DDFASGTSSWSTRLIHGGLRY 63
>GPDM_YEAST (P32191) Glycerol-3-phosphate dehydrogenase,
mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M)
Length = 649
Score = 44.3 bits (103), Expect = 7e-04
Identities = 25/89 (28%), Positives = 41/89 (45%), Gaps = 7/89 (7%)
Query: 41 RSTTQNDTNTTTSYP------VSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGR 94
R ND + +P VSR + D ++IG G G A A +G
Sbjct: 32 RPLVHNDPSYMVQFPTAAPPQVSRRDLLDRLAKTHQFDVLIIGGGATGTGCALDAATRGL 91
Query: 95 EVIVIESAPSFGTGTSSRNSEVVHAGIYY 123
V ++E F +GTSS++++++H G+ Y
Sbjct: 92 NVALVEKG-DFASGTSSKSTKMIHGGVRY 119
>GLPO_ENTCA (O86963) Alpha-glycerophosphate oxidase (EC 1.1.3.21)
(Glycerol-3-phosphate oxidase)
Length = 608
Score = 43.5 bits (101), Expect = 0.001
Identities = 21/52 (40%), Positives = 32/52 (61%), Gaps = 1/52 (1%)
Query: 72 DCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYY 123
D ++IG G+ G VA A G + +++E F GTSSR++++VH GI Y
Sbjct: 20 DVLIIGGGITGAGVAVQTAAAGMKTVLLEMQ-DFAEGTSSRSTKLVHGGIRY 70
>GLPO_STRPY (Q99YI8) Alpha-glycerophosphate oxidase (EC 1.1.3.21)
(Glycerol-3-phosphate oxidase)
Length = 612
Score = 43.1 bits (100), Expect = 0.002
Identities = 36/140 (25%), Positives = 63/140 (44%), Gaps = 13/140 (9%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGI-YYPHHSLK 129
+D ++IG G+ G VA A G + +IE F GTSSR++++VH G+ Y ++
Sbjct: 20 LDLLIIGGGITGAGVALQAAASGLDTGLIEMQ-DFAQGTSSRSTKLVHGGLRYLKQFDVE 78
Query: 130 AIFCVKGREMLYEYCAKHDIP---------HEQTGKLIVATRSSEIPKLSVILNHGIQNG 180
+ + + A H IP +++ G R L +L G+ N
Sbjct: 79 VVSDTVSERAVVQQIAPH-IPKPDPMLLPVYDEPGSTFSMFRLKVAMDLYDLL-AGVSNT 136
Query: 181 VDGLKMMDGVDAMKMEPELQ 200
K++ + +K EP+L+
Sbjct: 137 PAANKVLTKEEVLKREPDLK 156
>GLPO_STRPN (P35596) Alpha-glycerophosphate oxidase (EC 1.1.3.21)
(Glycerol-3-phosphate oxidase) (Exported protein 6)
Length = 608
Score = 43.1 bits (100), Expect = 0.002
Identities = 22/53 (41%), Positives = 32/53 (59%), Gaps = 1/53 (1%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYY 123
+D ++IG G+ G VA A G E +IE F GTSSR++++VH G+ Y
Sbjct: 20 LDLLIIGGGITGAGVALQAAASGLETGLIEMQ-DFAEGTSSRSTKLVHGGLRY 71
>GLPO_STRP3 (Q8K666) Alpha-glycerophosphate oxidase (EC 1.1.3.21)
(Glycerol-3-phosphate oxidase)
Length = 612
Score = 43.1 bits (100), Expect = 0.002
Identities = 36/140 (25%), Positives = 63/140 (44%), Gaps = 13/140 (9%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGI-YYPHHSLK 129
+D ++IG G+ G VA A G + +IE F GTSSR++++VH G+ Y ++
Sbjct: 20 LDLLIIGGGITGAGVALQAAASGLDTGLIEMQ-DFAQGTSSRSTKLVHGGLRYLKQFDVE 78
Query: 130 AIFCVKGREMLYEYCAKHDIP---------HEQTGKLIVATRSSEIPKLSVILNHGIQNG 180
+ + + A H IP +++ G R L +L G+ N
Sbjct: 79 VVSDTVSERAVVQQIAPH-IPKPDPMLLPVYDEPGSTFSMFRLKVAMDLYDLL-AGVSNT 136
Query: 181 VDGLKMMDGVDAMKMEPELQ 200
K++ + +K EP+L+
Sbjct: 137 PAANKVLTKEEVLKREPDLK 156
>GLPO_STRP8 (Q8NZX0) Alpha-glycerophosphate oxidase (EC 1.1.3.21)
(Glycerol-3-phosphate oxidase)
Length = 612
Score = 42.7 bits (99), Expect = 0.002
Identities = 36/140 (25%), Positives = 63/140 (44%), Gaps = 13/140 (9%)
Query: 71 VDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGI-YYPHHSLK 129
+D ++IG G+ G VA A G + +IE F GTSSR++++VH G+ Y ++
Sbjct: 20 LDLLIIGGGITGAGVALQAAASGLDTGLIEMQ-DFAQGTSSRSTKLVHGGLRYLKQFDVE 78
Query: 130 AIFCVKGREMLYEYCAKHDIP---------HEQTGKLIVATRSSEIPKLSVILNHGIQNG 180
+ + + A H IP +++ G R L +L G+ N
Sbjct: 79 VVSDTVSERAVVQQIAPH-IPKPDPMLLPVYDEPGSTFSMFRLKVAMDLYDLL-AGVSNM 136
Query: 181 VDGLKMMDGVDAMKMEPELQ 200
K++ + +K EP+L+
Sbjct: 137 PAANKVLTKEEVLKREPDLK 156
>YJ42_CHRVO (Q7NWN9) Hypothetical UPF0209 protein CV1942
Length = 660
Score = 42.4 bits (98), Expect = 0.003
Identities = 28/98 (28%), Positives = 48/98 (48%), Gaps = 15/98 (15%)
Query: 68 RERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNSEVVHAGIYYPH-- 125
RER +V+G G+ G A AR+LAL+G +V +IE P + S V++ + PH
Sbjct: 253 RER-SAIVVGGGLAGAASARSLALRGWQVTLIERMPQLASAASGNPQGVLYTKL-SPHLT 310
Query: 126 -----------HSLKAIFCVKGREMLYEYCAKHDIPHE 152
+SL+A+ C+ + ++ C + H+
Sbjct: 311 PLTRLVLSGYAYSLRALRCLLPEDGDWQACGVLQLAHD 348
>GPDM_SCHPO (O14400) Glycerol-3-phosphate dehydrogenase,
mitochondrial precursor (EC 1.1.99.5) (GPD-M) (GPDH-M)
Length = 649
Score = 42.0 bits (97), Expect = 0.003
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 4/78 (5%)
Query: 55 PVSRETTTSYSVPRERVDCVVIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRNS 114
P SRET + D ++IG G G VA + +G V ++E F + TSS+++
Sbjct: 52 PPSRETLLKNVEDISKFDVLIIGGGATGTGVAVDASTRGLNVCLLEKT-DFASETSSKST 110
Query: 115 EVVHAGIYYPHHSLKAIF 132
++ H G+ Y KA+F
Sbjct: 111 KMAHGGVRYLE---KAVF 125
>DAD2_RALSO (Q8XX54) D-amino acid dehydrogenase 2 small subunit (EC
1.4.99.1)
Length = 425
Score = 42.0 bits (97), Expect = 0.003
Identities = 20/39 (51%), Positives = 27/39 (68%)
Query: 75 VIGAGVVGIAVARALALKGREVIVIESAPSFGTGTSSRN 113
V+GAG+VGI+ A ALA +G +V ++E P G GTS N
Sbjct: 5 VVGAGIVGISTAYALAQEGHQVTLVERNPGPGEGTSYAN 43
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 56,300,919
Number of Sequences: 164201
Number of extensions: 2451421
Number of successful extensions: 7375
Number of sequences better than 10.0: 178
Number of HSP's better than 10.0 without gapping: 119
Number of HSP's successfully gapped in prelim test: 59
Number of HSP's that attempted gapping in prelim test: 7193
Number of HSP's gapped (non-prelim): 215
length of query: 470
length of database: 59,974,054
effective HSP length: 114
effective length of query: 356
effective length of database: 41,255,140
effective search space: 14686829840
effective search space used: 14686829840
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146910.5


