

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.4 + phase: 0
(321 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
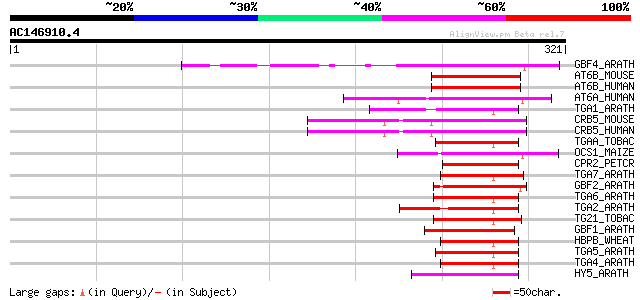
Score E
Sequences producing significant alignments: (bits) Value
GBF4_ARATH (P42777) G-box binding factor 4 (AtbZIP40) 74 4e-13
AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor AT... 51 4e-06
AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor AT... 51 4e-06
AT6A_HUMAN (P18850) Cyclic-AMP-dependent transcription factor AT... 50 8e-06
TGA1_ARATH (Q39237) Transcription factor TGA1 (DNA binding prote... 50 1e-05
CRB5_MOUSE (Q8K1L0) cAMP response element binding protein 5 (CRE... 49 1e-05
CRB5_HUMAN (Q02930) cAMP response element binding protein 5 (CRE... 49 1e-05
TGAA_TOBAC (P14232) TGACG-sequence specific DNA-binding protein ... 48 3e-05
OCS1_MAIZE (P24068) Ocs-element binding factor 1 (OCSBF-1) 48 3e-05
CPR2_PETCR (Q99090) Light-inducible protein CPRF-2 47 5e-05
TGA7_ARATH (Q93ZE2) Transcription factor TGA7 (AtbZIP50) 47 6e-05
GBF2_ARATH (P42775) G-box binding factor 2 (AtbZIP54) 47 6e-05
TGA6_ARATH (Q39140) Transcription factor TGA6 (AtbZIP45) 45 2e-04
TGA2_ARATH (P43273) Transcription factor TGA2 (HBP-1b homolog) (... 45 2e-04
TG21_TOBAC (O24160) TGACG-sequence specific DNA-binding protein ... 45 2e-04
GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41) 45 2e-04
HBPB_WHEAT (P23923) Transcription factor HBP-1b(c38) 45 2e-04
TGA5_ARATH (Q39163) Transcription factor TGA5 (Ocs-element bindi... 45 3e-04
TGA4_ARATH (Q39162) Transcription factor TGA4 (Ocs-element bindi... 45 3e-04
HY5_ARATH (O24646) Transcription factor HY5 (LONG HYPOCOL5 prote... 44 4e-04
>GBF4_ARATH (P42777) G-box binding factor 4 (AtbZIP40)
Length = 270
Score = 74.3 bits (181), Expect = 4e-13
Identities = 67/231 (29%), Positives = 108/231 (46%), Gaps = 60/231 (25%)
Query: 100 KTVDEVWKDM-QGKKRGVDRDRKSREKQQTLGEMTLEDFLVKAGVVGESFHGKESGLLRV 158
K+VD+VWK++ G+++ + ++++ MTLEDFL KA + E +
Sbjct: 85 KSVDDVWKEIVSGEQKTI-----MMKEEEPEDIMTLEDFLAKAEM-------DEGASDEI 132
Query: 159 DSNEDSRQKVSHGLHWMQYPVHSVQQQQHQYEKHTMPGFAAVHAIQQPFQVAGNQALDAA 218
D + + + G + +P+ Q+H FQ+
Sbjct: 133 DVKIPTERLNNDGSYTFDFPM-----QRHS-----------------SFQM--------- 161
Query: 219 ISPSSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELE 278
+ G++ T G++ ++K +RQKRMIKNRESAARSR R+QAY ELE
Sbjct: 162 -----VEGSMGGGVTRGKRGRVMMEAMDKAAAQRQKRMIKNRESAARSRERKQAYQVELE 216
Query: 279 LKVSRLEEENERLRRQNE----------MEKEVPTAPPPEPKNQ-LRRTNS 318
++LEEENE+L ++ E ME +P P P ++ L R++S
Sbjct: 217 TLAAKLEEENEQLLKEIEESTKERYKKLMEVLIPVDEKPRPPSRPLSRSHS 267
>AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp)
Length = 699
Score = 50.8 bits (120), Expect = 4e-06
Identities = 23/51 (45%), Positives = 38/51 (74%)
Query: 245 VEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQN 295
V+ + +RQ+RMIKNRESA +SR +++ Y Q LE ++ + +N++LRR+N
Sbjct: 318 VDAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQLRREN 368
>AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp) (G13 protein)
Length = 703
Score = 50.8 bits (120), Expect = 4e-06
Identities = 23/51 (45%), Positives = 38/51 (74%)
Query: 245 VEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQN 295
V+ + +RQ+RMIKNRESA +SR +++ Y Q LE ++ + +N++LRR+N
Sbjct: 321 VDAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQLRREN 371
>AT6A_HUMAN (P18850) Cyclic-AMP-dependent transcription factor ATF-6
alpha (Activating transcription factor 6 alpha)
(ATF6-alpha)
Length = 670
Score = 50.1 bits (118), Expect = 8e-06
Identities = 38/131 (29%), Positives = 66/131 (50%), Gaps = 12/131 (9%)
Query: 194 MPGFAAVHAIQQPFQVAGNQALDAAISPSS---LMGTLSDTQTLGRKRVASGIVVEKTVE 250
+P V A+ N ++ +PS+ + G LS T+ + + + + + + V
Sbjct: 249 LPSAQPVLAVAGGVTQLPNHVVNVVPAPSANSPVNGKLSVTKPVLQSTMRN-VGSDIAVL 307
Query: 251 RRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQN--------EMEKEVP 302
RRQ+RMIKNRESA +SR +++ Y LE ++ ENE+L+++N E+ E
Sbjct: 308 RRQQRMIKNRESACQSRKKKKEYMLGLEARLKAALSENEQLKKENGTLKRQLDEVVSENQ 367
Query: 303 TAPPPEPKNQL 313
P PK ++
Sbjct: 368 RLKVPSPKRRV 378
>TGA1_ARATH (Q39237) Transcription factor TGA1 (DNA binding protein
TGA1a-like protein) (AtbZIP47)
Length = 368
Score = 49.7 bits (117), Expect = 1e-05
Identities = 30/89 (33%), Positives = 51/89 (56%), Gaps = 8/89 (8%)
Query: 209 VAGNQALDAAISPSSLMGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRA 268
+ NQ LD +S + GT ++ S + ++ Q+R+ +NRE+A +SR
Sbjct: 47 IPNNQKLDNNVSEDTSHGTAGTPHMFDQEASTS-----RHPDKIQRRLAQNREAARKSRL 101
Query: 269 RRQAYTQELE---LKVSRLEEENERLRRQ 294
R++AY Q+LE LK+ +LE+E +R R+Q
Sbjct: 102 RKKAYVQQLETSRLKLIQLEQELDRARQQ 130
>CRB5_MOUSE (Q8K1L0) cAMP response element binding protein 5
(CRE-BPa) (Fragment)
Length = 353
Score = 49.3 bits (116), Expect = 1e-05
Identities = 38/136 (27%), Positives = 62/136 (44%), Gaps = 11/136 (8%)
Query: 173 HWMQYPVHSVQQQQHQYEKHTMP-GFAAVHAIQQPFQVAGNQAL----DAAISPSSLMGT 227
H Q+P H Q H + H + +HA Q + + L A +SP++
Sbjct: 137 HQHQHPAHHPHPQPHHQQNHPHHHSHSHLHAHPAHHQTSPHPPLHTGNQAQVSPATQQ-- 194
Query: 228 LSDTQTLGRKRVASG----IVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSR 283
+ TQT+ + G +V E ERR+K + +NR +A R R +R+ + LE K
Sbjct: 195 MQPTQTIQPPQPTGGRRRRVVDEDPDERRRKFLERNRAAATRCRQKRKVWVMSLEKKAEE 254
Query: 284 LEEENERLRRQNEMEK 299
L + N +L+ + M K
Sbjct: 255 LTQTNMQLQNEVSMLK 270
>CRB5_HUMAN (Q02930) cAMP response element binding protein 5
(CRE-BPa)
Length = 508
Score = 49.3 bits (116), Expect = 1e-05
Identities = 38/136 (27%), Positives = 62/136 (44%), Gaps = 11/136 (8%)
Query: 173 HWMQYPVHSVQQQQHQYEKHTMP-GFAAVHAIQQPFQVAGNQAL----DAAISPSSLMGT 227
H Q+P H Q H + H + +HA Q + + L A +SP++
Sbjct: 292 HQHQHPAHHPHPQPHHQQNHPHHHSHSHLHAHPAHHQTSPHPPLHTGNQAQVSPATQQ-- 349
Query: 228 LSDTQTLGRKRVASG----IVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSR 283
+ TQT+ + G +V E ERR+K + +NR +A R R +R+ + LE K
Sbjct: 350 MQPTQTIQPPQPTGGRRRRVVDEDPDERRRKFLERNRAAATRCRQKRKVWVMSLEKKAEE 409
Query: 284 LEEENERLRRQNEMEK 299
L + N +L+ + M K
Sbjct: 410 LTQTNMQLQNEVSMLK 425
>TGAA_TOBAC (P14232) TGACG-sequence specific DNA-binding protein
TGA-1A (TGA1a) (ASF-1 protein)
Length = 359
Score = 48.1 bits (113), Expect = 3e-05
Identities = 25/51 (49%), Positives = 38/51 (74%), Gaps = 3/51 (5%)
Query: 247 KTVERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQ 294
K VE+ +R+ +NRE+A +SR R++AY Q+LE LK+ +LE+E ER R+Q
Sbjct: 70 KPVEKVLRRLAQNREAARKSRLRKKAYVQQLENSKLKLIQLEQELERARKQ 120
>OCS1_MAIZE (P24068) Ocs-element binding factor 1 (OCSBF-1)
Length = 151
Score = 48.1 bits (113), Expect = 3e-05
Identities = 36/103 (34%), Positives = 51/103 (48%), Gaps = 11/103 (10%)
Query: 225 MGTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRL 284
M + S + T GR + G T RR+KR + NRESA RSR R+Q + EL +V+RL
Sbjct: 1 MSSSSLSPTAGRTSGSDGDSAADT-HRREKRRLSNRESARRSRLRKQQHLDELVQEVARL 59
Query: 285 EEENERLRRQN----------EMEKEVPTAPPPEPKNQLRRTN 317
+ +N R+ + E E V A E ++LR N
Sbjct: 60 QADNARVAARARDIASQYTRVEQENTVLRARAAELGDRLRSVN 102
>CPR2_PETCR (Q99090) Light-inducible protein CPRF-2
Length = 401
Score = 47.4 bits (111), Expect = 5e-05
Identities = 23/44 (52%), Positives = 32/44 (72%)
Query: 251 RRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ 294
+R +RM+ NRESA RSR R+QA+ ELE +VS+L EN L ++
Sbjct: 200 KRVRRMLSNRESARRSRRRKQAHMTELETQVSQLRVENSSLLKR 243
>TGA7_ARATH (Q93ZE2) Transcription factor TGA7 (AtbZIP50)
Length = 368
Score = 47.0 bits (110), Expect = 6e-05
Identities = 21/51 (41%), Positives = 40/51 (78%), Gaps = 3/51 (5%)
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQNEM 297
++ ++R+ +NRE+A +SR R++AY Q+LE LK+S+LE+E E++++Q +
Sbjct: 92 DKMKRRLAQNREAARKSRLRKKAYVQQLEESRLKLSQLEQELEKVKQQGHL 142
>GBF2_ARATH (P42775) G-box binding factor 2 (AtbZIP54)
Length = 360
Score = 47.0 bits (110), Expect = 6e-05
Identities = 30/59 (50%), Positives = 37/59 (61%), Gaps = 6/59 (10%)
Query: 246 EKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLRRQ-----NEMEK 299
EK V +R+KR NRESA RSR R+QA T++L +KV L EN LR + NE EK
Sbjct: 247 EKEV-KREKRKQSNRESARRSRLRKQAETEQLSVKVDALVAENMSLRSKLGQLNNESEK 304
>TGA6_ARATH (Q39140) Transcription factor TGA6 (AtbZIP45)
Length = 330
Score = 45.4 bits (106), Expect = 2e-04
Identities = 22/52 (42%), Positives = 39/52 (74%), Gaps = 3/52 (5%)
Query: 246 EKTVERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQ 294
+K ++ +R+ +NRE+A +SR R++AY Q+LE LK+++LE+E +R R+Q
Sbjct: 41 DKLDQKTLRRLAQNREAARKSRLRKKAYVQQLENSRLKLTQLEQELQRARQQ 92
>TGA2_ARATH (P43273) Transcription factor TGA2 (HBP-1b homolog)
(AHBP-1b) (AtbZIP20)
Length = 330
Score = 45.4 bits (106), Expect = 2e-04
Identities = 27/72 (37%), Positives = 46/72 (63%), Gaps = 7/72 (9%)
Query: 226 GTLSDTQTLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELE---LKVS 282
G L +T + G + +KT+ +R+ +NRE+A +SR R++AY Q+LE LK++
Sbjct: 25 GALVNTAASDSSDRSKGKMDQKTL----RRLAQNREAARKSRLRKKAYVQQLENSRLKLT 80
Query: 283 RLEEENERLRRQ 294
+LE+E +R R+Q
Sbjct: 81 QLEQELQRARQQ 92
>TG21_TOBAC (O24160) TGACG-sequence specific DNA-binding protein
TGA-2.1 (TGA2.1)
Length = 456
Score = 45.4 bits (106), Expect = 2e-04
Identities = 23/55 (41%), Positives = 42/55 (75%), Gaps = 4/55 (7%)
Query: 246 EKTVERRQ-KRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQNE 296
EK ++++ +R+ +NRE+A +SR R++AY Q+LE LK+S+LE++ +R R+Q +
Sbjct: 162 EKVLDQKTLRRLAQNREAARKSRLRKKAYVQQLENSRLKLSQLEQDLQRARQQGK 216
>GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41)
Length = 315
Score = 45.4 bits (106), Expect = 2e-04
Identities = 25/53 (47%), Positives = 35/53 (65%), Gaps = 1/53 (1%)
Query: 241 SGIVVEKTVE-RRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLR 292
+G+ V+ E +RQKR NRESA RSR R+QA ++L+ +V L EN+ LR
Sbjct: 213 AGVPVKDERELKRQKRKQSNRESARRSRLRKQAECEQLQQRVESLSNENQSLR 265
>HBPB_WHEAT (P23923) Transcription factor HBP-1b(c38)
Length = 332
Score = 45.1 bits (105), Expect = 2e-04
Identities = 21/48 (43%), Positives = 37/48 (76%), Gaps = 3/48 (6%)
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQ 294
++ +R+ +NRE+A +SR R++AY Q+LE LK+++LE+E +R R+Q
Sbjct: 45 QKTMRRLAQNREAARKSRLRKKAYVQQLENSRLKLTQLEQELQRARQQ 92
>TGA5_ARATH (Q39163) Transcription factor TGA5 (Ocs-element binding
factor 5) (OBF5) (AtbZIP26)
Length = 330
Score = 44.7 bits (104), Expect = 3e-04
Identities = 22/51 (43%), Positives = 38/51 (74%), Gaps = 3/51 (5%)
Query: 247 KTVERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQ 294
K ++ +R+ +NRE+A +SR R++AY Q+LE LK+++LE+E +R R+Q
Sbjct: 42 KMDQKTLRRLAQNREAARKSRLRKKAYVQQLENSRLKLTQLEQELQRARQQ 92
>TGA4_ARATH (Q39162) Transcription factor TGA4 (Ocs-element binding
factor 4) (OBF4) (AtbZIP57)
Length = 364
Score = 44.7 bits (104), Expect = 3e-04
Identities = 22/48 (45%), Positives = 36/48 (74%), Gaps = 3/48 (6%)
Query: 250 ERRQKRMIKNRESAARSRARRQAYTQELE---LKVSRLEEENERLRRQ 294
++ Q+R+ +NRE+A +SR R++AY Q+LE LK+ LE+E +R R+Q
Sbjct: 79 DKIQRRLAQNREAARKSRLRKKAYVQQLETSRLKLIHLEQELDRARQQ 126
>HY5_ARATH (O24646) Transcription factor HY5 (LONG HYPOCOL5 protein)
(AtbZIP56)
Length = 168
Score = 44.3 bits (103), Expect = 4e-04
Identities = 22/62 (35%), Positives = 36/62 (57%)
Query: 233 TLGRKRVASGIVVEKTVERRQKRMIKNRESAARSRARRQAYTQELELKVSRLEEENERLR 292
T+G + G + +R KR+++NR SA ++R R++AY ELE +V LE +N L
Sbjct: 72 TVGESQRKRGRTPAEKENKRLKRLLRNRVSAQQARERKKAYLSELENRVKDLENKNSELE 131
Query: 293 RQ 294
+
Sbjct: 132 ER 133
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.310 0.127 0.349
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,618,180
Number of Sequences: 164201
Number of extensions: 1521881
Number of successful extensions: 7154
Number of sequences better than 10.0: 263
Number of HSP's better than 10.0 without gapping: 147
Number of HSP's successfully gapped in prelim test: 118
Number of HSP's that attempted gapping in prelim test: 6638
Number of HSP's gapped (non-prelim): 615
length of query: 321
length of database: 59,974,054
effective HSP length: 110
effective length of query: 211
effective length of database: 41,911,944
effective search space: 8843420184
effective search space used: 8843420184
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC146910.4


