

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146910.2 + phase: 1 /pseudo/partial
(757 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
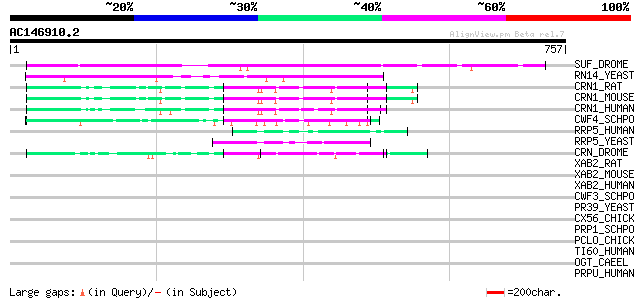
SUF_DROME (P25991) Suppressor of forked protein 333 8e-91
RN14_YEAST (P25298) mRNA 3'-end processing protein RNA14 157 8e-38
CRN1_RAT (P63155) Crooked neck-like protein 1 (Crooked neck homo... 59 5e-08
CRN1_MOUSE (P63154) Crooked neck-like protein 1 (Crooked neck ho... 59 5e-08
CRN1_HUMAN (Q9BZJ0) Crooked neck-like protein 1 (Crooked neck ho... 59 5e-08
CWF4_SCHPO (P87312) Cell cycle control protein cwf4 58 8e-08
RRP5_HUMAN (Q14690) RRP5 protein homolog (Programmed cell death ... 52 6e-06
RRP5_YEAST (Q05022) rRNA biogenesis protein RRP5 50 2e-05
CRN_DROME (P17886) Crooked neck protein 48 8e-05
XAB2_RAT (Q99PK0) XPA-binding protein 2 (Adapter protein ATH-55) 44 0.001
XAB2_MOUSE (Q9DCD2) XPA-binding protein 2 44 0.001
XAB2_HUMAN (Q9HCS7) XPA-binding protein 2 (HCNP protein) (PP3898) 44 0.001
CWF3_SCHPO (Q9P7R9) Cell cycle control protein cwf3 39 0.051
PR39_YEAST (P39682) Pre-mRNA processing protein PRP39 37 0.25
CX56_CHICK (P29415) Gap junction Cx56 protein (Connexin 56) 36 0.33
PRP1_SCHPO (Q12381) Pre-mRNA splicing factor prp1 35 0.56
PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment) 35 0.73
TI60_HUMAN (Q92993) Histone acetyltransferase HTATIP (EC 2.3.1.4... 34 1.3
OGT_CAEEL (O18158) UDP-N-acetylglucosamine--peptide N-acetylgluc... 34 1.3
PRPU_HUMAN (O94906) U5 snRNP-associated 102 kDa protein (U5-102 ... 34 1.6
>SUF_DROME (P25991) Suppressor of forked protein
Length = 733
Score = 333 bits (855), Expect = 8e-91
Identities = 221/733 (30%), Positives = 363/733 (49%), Gaps = 76/733 (10%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
I E +YE L+ ++PT A++WK Y+E M + ++++F RCL+ L + LW+ Y+
Sbjct: 48 IHEVRSLYESLVNVFPTTARYWKLYIEMEMRSRYYERVEKLFQRCLVKILNIDLWKLYLT 107
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
++++ +E+ +A++F L +G D+ S +W +YI FL+ + A E ++
Sbjct: 108 YVKETKSGLSTH-KEKMAQAYDFALEKIGMDLHSFSIWQDYIYFLRGVEAVGNYAENQKI 166
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
T VR+VYQ+A++TP IEQLWKDY +FE +++ +++ + E Y +AR V +E +
Sbjct: 167 TAVRRVYQKAVVTPIVGIEQLWKDYIAFEQNINPIISEKMSLERSKDYMNARRVAKELEY 226
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
++ N+ AVPPT + + K + L K +++
Sbjct: 227 HTKGLNRNLPAVPPTLTKEEVKQVELWKRFITY--------------------------- 259
Query: 263 RNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWH-------AK 315
E S + D A +RV+F EQCL+ L H+P VW+ + + +
Sbjct: 260 --------EKSNPLRTEDTALVTRRVMFATEQCLLVLTHHPAVWHQASQFLDTSARVLTE 311
Query: 316 AGSIDAA-------IKVFQRSLKA-LPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDS 367
G + AA + +RS+ L + +L +AYA+ EE R + +Y LL
Sbjct: 312 KGDVQAAKIFADECANILERSINGVLNRNALLYFAYADFEEGRLKYEKVHTMYNKLLQLP 371
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ L ++Q+++F RR EG++ AR F AR+ YH++VA A + + KD ++A
Sbjct: 372 DIDPTLVYVQYMKFARRAEGIKSARSIFKKAREDVRSRYHIFVAAALMEYYCSKDKEIAF 431
Query: 428 NVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS--LPLEDSVEVWKRFVK 485
+FE GLK F P Y++ Y D+L LN+D N R LFER LSS L SVEVW RF++
Sbjct: 432 RIFELGLKRFGGSPEYVMCYIDYLSHLNEDNNTRVLFERVLSSGGLSPHKSVEVWNRFLE 491
Query: 486 FEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQ 545
FE GDL+S++KVE+RR F E +V RY F+DL+PC+S +L ++
Sbjct: 492 FESNIGDLSSIVKVERRRSAVF-ENLKEYEGKETAQLVDRYKFLDLYPCTSTELKSIGYA 550
Query: 546 EWLVKNTKKVEKSIMLNGTTFIDKGPVASISTTSSKVVYPDTSKMLIYDPK--HNPGTGA 603
E + KV G + G V + S + + PD S+M+ + P+ +PG
Sbjct: 551 ENVGIILNKVG-----GGAQSQNTGEVETDSEATPPLPRPDFSQMIPFKPRPCAHPGAHP 605
Query: 604 AGTNAFDEILKATPPALVAFLANLP---SVDGPTPNVDIVLSICLQSDLPTGQSVKVGIP 660
F + PPAL A A LP S GP +V+++ I ++ +LP G
Sbjct: 606 LAGGVFPQ-----PPALAALCATLPPPNSFRGPFVSVELLFDIFMRLNLPDSAPQPNGDN 660
Query: 661 SQLPA--GPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
P A + + +S VQ ++ + P R R+ L ++ D + + P
Sbjct: 661 ELSPKIFDLAKSVHWIVDTSTYTGVQHSVTAVPPRR-----RRLLPGGDDSDDELQTAVP 715
Query: 719 LPQDAFRIRQFQK 731
D +R+RQ ++
Sbjct: 716 PSHDIYRLRQLKR 728
>RN14_YEAST (P25298) mRNA 3'-end processing protein RNA14
Length = 677
Score = 157 bits (398), Expect = 8e-38
Identities = 125/535 (23%), Positives = 235/535 (43%), Gaps = 84/535 (15%)
Query: 22 SIAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQ---VPLWR 78
S A+ +YEQ +P + W ++ +A + + +++I ++CL L+ + LW
Sbjct: 57 SYAKVREVYEQFHNTFPFYSPAWTLQLKGELARDEFETVEKILAQCLSGKLENNDLSLWS 116
Query: 79 CYIRFIRKVND--KKGAEGQEETKKAFEFMLSYVGS-DIASGPVWMEYIAFLKSLPAAHP 135
Y+ +IR+ N+ G E + KAF+ ++ + S W EY+ FL+ +
Sbjct: 117 TYLDYIRRKNNLITGGQEARAVIVKAFQLVMQKCAIFEPKSSSFWNEYLNFLEQWKPFNK 176
Query: 136 QEETHRMTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARA 195
EE R+ ++R+ Y++ + P ++E++W Y +E ++ A+ I E +Y AR+
Sbjct: 177 WEEQQRIDMLREFYKKMLCVPFDNLEKMWNRYTQWEQEINSLTARKFIGELSAEYMKARS 236
Query: 196 VYRE---------RKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWN 246
+Y+E R + N +P G+ ++ + L +++N + W
Sbjct: 237 LYQEWLNVTNGLKRASPINLRTANKKNIPQPGTSDSN---------IQQLQIWLNWIKWE 287
Query: 247 DNHFPPKSLCLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVW 306
N + + E+ ++R+ + Y+Q + Y+ ++W
Sbjct: 288 RE---------------NKLMLSEDML-----------SQRISYVYKQGIQYMIFSAEMW 321
Query: 307 YDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEE---------------SRG 351
YDY+ + ++ + +L A PDS L + +E E ++
Sbjct: 322 YDYSMYISENSDRQ---NILYTALLANPDSPSLTFKLSECYELDNDSESVSNCFDKCTQT 378
Query: 352 AIQAAKKIYENLLGDSENAT--------------ALAHIQFIRFLRRTEGVEPARKYFLD 397
+ KKI ++ +N T ++ ++R G+ AR F
Sbjct: 379 LLSQYKKIASDVNSGEDNNTEYEQELLYKQREKLTFVFCVYMNTMKRISGLSAARTVFGK 438
Query: 398 ARKSPSC-TYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLND 456
RK T+ VYV A + F D K A V E GLK+F ++ VYI +Y DFLI LN
Sbjct: 439 CRKLKRILTHDVYVENAYLEFQNQNDYKTAFKVLELGLKYFQNDGVYINKYLDFLIFLNK 498
Query: 457 DQNIRALFERALSSL-PLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
D I+ LFE ++ + L E++K+ + +E +G+L ++ +E+R E F +E
Sbjct: 499 DSQIKTLFETSVEKVQDLTQLKEIYKKMISYESKFGNLNNVYSLEKRFFERFPQE 553
>CRN1_RAT (P63155) Crooked neck-like protein 1 (Crooked neck
homolog) (Crooked neck protein)
Length = 690
Score = 58.9 bits (141), Expect = 5e-08
Identities = 52/229 (22%), Positives = 100/229 (42%), Gaps = 14/229 (6%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRG 351
YE+ L Y +W YA K ++ A ++ R++ LP Y Y +EE G
Sbjct: 104 YERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYTYMEEMLG 163
Query: 352 AIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVA 411
+ A++++E + A +I F R + VE AR + + +
Sbjct: 164 NVAGARQVFERWM--EWQPEEQAWHSYINFELRYKEVERARTIY----ERFVLVHPAVKN 217
Query: 412 YASVAFCLDKDPKMAH--NVFEAGLKHF----MHEPVYILEYADFLIRLNDDQNIRALFE 465
+ A +K AH V+E ++ F M E +Y+ +A F + + +R +++
Sbjct: 218 WIKYARFEEKHAYFAHARKVYERAVEFFGDEHMDEHLYV-AFAKFEENQKEFERVRVIYK 276
Query: 466 RALSSLPLEDSVEVWKRFVKFEQTYGDLASMLK-VEQRRKEAFGEEATA 513
AL + +++ E++K + FE+ +GD + + +R+ + EE A
Sbjct: 277 YALDRISKQEAQELFKNYTIFEKKFGDRRGIEDIIVSKRRFQYEEEVKA 325
Score = 50.4 bits (119), Expect = 2e-05
Identities = 101/488 (20%), Positives = 177/488 (35%), Gaps = 82/488 (16%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQV-PLWRCYI 81
I A IYE+ L + W +Y E M + + I+ R + +V W Y
Sbjct: 97 IQRARSIYERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYT 156
Query: 82 RFIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHR 141
+ + GA ++ FE + + + A W YI F +E R
Sbjct: 157 YMEEMLGNVAGA------RQVFERWMEWQPEEQA----WHSYINFELRY------KEVER 200
Query: 142 MTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERK 201
R +Y+R ++ H + W Y FE E + AR VY
Sbjct: 201 ---ARTIYERFVLV--HPAVKNWIKYARFE-------------EKHAYFAHARKVYERAV 242
Query: 202 KFF-----DEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVF-VNKLFWNDNHFPPKSL 255
+FF DE + A + + + KY L +S +LF N F K
Sbjct: 243 EFFGDEHMDEHLYVAFAKFEENQKEFERVRVIYKYALDRISKQEAQELFKNYTIFEKK-- 300
Query: 256 CLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAK 315
R ++ DI S +R F YE+ + H D W+DY
Sbjct: 301 ----FGDRRGIE------------DIIVSKRR--FQYEEEVKANPHNYDAWFDYLRLVES 342
Query: 316 AGSIDAAIKVFQRSLKALPDSEMLRY------------AYAELEESRGAIQAAKKIYE-- 361
D +V++R++ +P + R+ Y ELE + +++Y+
Sbjct: 343 DAEADTVREVYERAIANVPPIQEKRHWKRYIYLWVNYALYEELEAKDP--ERTRQVYQAS 400
Query: 362 -NLLGDSENATALAHIQFIRFLRRTEGVEPARKYF-LDARKSPSCTYHVYVAYASVAFCL 419
L+ + A + + +F R + + AR+ K P ++ Y + L
Sbjct: 401 LELIPHKKFTFAKMWLYYAQFEIRQKNLPFARRALGTSIGKCPK--NKLFKGYIELELQL 458
Query: 420 DKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEV 479
++ ++E L+ +++A+ L D + RA++E A+S L+ +
Sbjct: 459 -REFDRCRKLYEKFLEFGPENCTSWIKFAELETILGDIERARAIYELAISQPRLDMPEVL 517
Query: 480 WKRFVKFE 487
WK ++ FE
Sbjct: 518 WKSYIDFE 525
Score = 49.3 bits (116), Expect = 4e-05
Identities = 110/552 (19%), Positives = 212/552 (37%), Gaps = 72/552 (13%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
+ A I+++ + P +FW +Y + N +Q+F R + + W YI
Sbjct: 131 VNHARNIWDRAITTLPRVNQFWYKYTYMEEMLGNVAGARQVFERWMEWQPEEQAWHSYIN 190
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
F + + + A E F+L + + W++Y F EE H
Sbjct: 191 FELRYKEVERARTIYE-----RFVLVH-----PAVKNWIKYARF----------EEKHAY 230
Query: 143 TV-VRKVYQRAI--ITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRE 199
RKVY+RA+ H E L+ + FE + QK + + Y KY R +E
Sbjct: 231 FAHARKVYERAVEFFGDEHMDEHLYVAFAKFEEN--QKEFERVRVIY--KYALDRISKQE 286
Query: 200 RKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFL 259
++ F + G + + + + K + ++ N +++ L L
Sbjct: 287 AQELFKNYT---IFEKKFGDRRGIEDIIVSKRRFQ----YEEEVKANPHNYDAWFDYLRL 339
Query: 260 LTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHA-KAGS 318
+ D + E + A +N I Y+Y +W +YA + +A
Sbjct: 340 VESDAEADTVREV------YERAIANVPPIQEKRHWKRYIY----LWVNYALYEELEAKD 389
Query: 319 IDAAIKVFQRSLKALPDSEM----LRYAYAELEESRGAIQAAKKIYENLLGDS-ENATAL 373
+ +V+Q SL+ +P + + YA+ E + + A++ +G +N
Sbjct: 390 PERTRQVYQASLELIPHKKFTFAKMWLYYAQFEIRQKNLPFARRALGTSIGKCPKNKLFK 449
Query: 374 AHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAG 433
+I+ LR + + FL+ +CT ++ +A + L D + A ++E
Sbjct: 450 GYIELELQLREFDRCRKLYEKFLEFGPE-NCTS--WIKFAELETILG-DIERARAIYELA 505
Query: 434 LKHFMHEPVYIL--EYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYG 491
+ + +L Y DF I + + R L+ + L V+VW F +FE + G
Sbjct: 506 ISQPRLDMPEVLWKSYIDFEIEQEETERTRNLYRQLLQRT---QHVKVWISFAQFELSSG 562
Query: 492 DLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWL--- 548
S+ K Q +EA + + ++ R ++ W ++ +S +E +
Sbjct: 563 KEGSVAKCRQIYEEA------NKTMRNCEEKEERLMLLESWRSFEDEFGTVSDKERVDKL 616
Query: 549 ----VKNTKKVE 556
VK +KV+
Sbjct: 617 MPEKVKKRRKVQ 628
Score = 35.0 bits (79), Expect = 0.73
Identities = 28/119 (23%), Positives = 56/119 (46%), Gaps = 3/119 (2%)
Query: 392 RKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFL 451
RK F D + ++ YA L K+ + A +++E L L+YA+
Sbjct: 67 RKTFEDNIRKNRTVISNWIKYAQWEESL-KEIQRARSIYERALDVDYRNITLWLKYAEME 125
Query: 452 IRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
++ + R +++RA+++LP + + W ++ E+ G++A +V +R E EE
Sbjct: 126 MKNRQVNHARNIWDRAITTLPRVN--QFWYKYTYMEEMLGNVAGARQVFERWMEWQPEE 182
>CRN1_MOUSE (P63154) Crooked neck-like protein 1 (Crooked neck
homolog)
Length = 690
Score = 58.9 bits (141), Expect = 5e-08
Identities = 52/229 (22%), Positives = 100/229 (42%), Gaps = 14/229 (6%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRG 351
YE+ L Y +W YA K ++ A ++ R++ LP Y Y +EE G
Sbjct: 104 YERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYTYMEEMLG 163
Query: 352 AIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVA 411
+ A++++E + A +I F R + VE AR + + +
Sbjct: 164 NVAGARQVFERWM--EWQPEEQAWHSYINFELRYKEVERARTIY----ERFVLVHPAVKN 217
Query: 412 YASVAFCLDKDPKMAH--NVFEAGLKHF----MHEPVYILEYADFLIRLNDDQNIRALFE 465
+ A +K AH V+E ++ F M E +Y+ +A F + + +R +++
Sbjct: 218 WIKYARFEEKHAYFAHARKVYERAVEFFGDEHMDEHLYV-AFAKFEENQKEFERVRVIYK 276
Query: 466 RALSSLPLEDSVEVWKRFVKFEQTYGDLASMLK-VEQRRKEAFGEEATA 513
AL + +++ E++K + FE+ +GD + + +R+ + EE A
Sbjct: 277 YALDRISKQEAQELFKNYTIFEKKFGDRRGIEDIIVSKRRFQYEEEVKA 325
Score = 50.4 bits (119), Expect = 2e-05
Identities = 101/488 (20%), Positives = 177/488 (35%), Gaps = 82/488 (16%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQV-PLWRCYI 81
I A IYE+ L + W +Y E M + + I+ R + +V W Y
Sbjct: 97 IQRARSIYERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYT 156
Query: 82 RFIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHR 141
+ + GA ++ FE + + + A W YI F +E R
Sbjct: 157 YMEEMLGNVAGA------RQVFERWMEWQPEEQA----WHSYINFELRY------KEVER 200
Query: 142 MTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERK 201
R +Y+R ++ H + W Y FE E + AR VY
Sbjct: 201 ---ARTIYERFVLV--HPAVKNWIKYARFE-------------EKHAYFAHARKVYERAV 242
Query: 202 KFF-----DEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVF-VNKLFWNDNHFPPKSL 255
+FF DE + A + + + KY L +S +LF N F K
Sbjct: 243 EFFGDEHMDEHLYVAFAKFEENQKEFERVRVIYKYALDRISKQEAQELFKNYTIFEKK-- 300
Query: 256 CLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAK 315
R ++ DI S +R F YE+ + H D W+DY
Sbjct: 301 ----FGDRRGIE------------DIIVSKRR--FQYEEEVKANPHNYDAWFDYLRLVES 342
Query: 316 AGSIDAAIKVFQRSLKALPDSEMLRY------------AYAELEESRGAIQAAKKIYE-- 361
D +V++R++ +P + R+ Y ELE + +++Y+
Sbjct: 343 DAEADTVREVYERAIANVPPIQEKRHWKRYIYLWVNYALYEELEAKDP--ERTRQVYQAS 400
Query: 362 -NLLGDSENATALAHIQFIRFLRRTEGVEPARKYF-LDARKSPSCTYHVYVAYASVAFCL 419
L+ + A + + +F R + + AR+ K P ++ Y + L
Sbjct: 401 LELIPHKKFTFAKMWLYYAQFEIRQKNLPFARRALGTSIGKCPK--NKLFKGYIELELQL 458
Query: 420 DKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEV 479
++ ++E L+ +++A+ L D + RA++E A+S L+ +
Sbjct: 459 -REFDRCRKLYEKFLEFGPENCTSWIKFAELETILGDIERARAIYELAISQPRLDMPEVL 517
Query: 480 WKRFVKFE 487
WK ++ FE
Sbjct: 518 WKSYIDFE 525
Score = 49.3 bits (116), Expect = 4e-05
Identities = 110/552 (19%), Positives = 212/552 (37%), Gaps = 72/552 (13%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
+ A I+++ + P +FW +Y + N +Q+F R + + W YI
Sbjct: 131 VNHARNIWDRAITTLPRVNQFWYKYTYMEEMLGNVAGARQVFERWMEWQPEEQAWHSYIN 190
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
F + + + A E F+L + + W++Y F EE H
Sbjct: 191 FELRYKEVERARTIYE-----RFVLVH-----PAVKNWIKYARF----------EEKHAY 230
Query: 143 TV-VRKVYQRAI--ITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRE 199
RKVY+RA+ H E L+ + FE + QK + + Y KY R +E
Sbjct: 231 FAHARKVYERAVEFFGDEHMDEHLYVAFAKFEEN--QKEFERVRVIY--KYALDRISKQE 286
Query: 200 RKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFL 259
++ F + G + + + + K + ++ N +++ L L
Sbjct: 287 AQELFKNYT---IFEKKFGDRRGIEDIIVSKRRFQ----YEEEVKANPHNYDAWFDYLRL 339
Query: 260 LTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHA-KAGS 318
+ D + E + A +N I Y+Y +W +YA + +A
Sbjct: 340 VESDAEADTVREV------YERAIANVPPIQEKRHWKRYIY----LWVNYALYEELEAKD 389
Query: 319 IDAAIKVFQRSLKALPDSEM----LRYAYAELEESRGAIQAAKKIYENLLGDS-ENATAL 373
+ +V+Q SL+ +P + + YA+ E + + A++ +G +N
Sbjct: 390 PERTRQVYQASLELIPHKKFTFAKMWLYYAQFEIRQKNLPFARRALGTSIGKCPKNKLFK 449
Query: 374 AHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAG 433
+I+ LR + + FL+ +CT ++ +A + L D + A ++E
Sbjct: 450 GYIELELQLREFDRCRKLYEKFLEFGPE-NCTS--WIKFAELETILG-DIERARAIYELA 505
Query: 434 LKHFMHEPVYIL--EYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYG 491
+ + +L Y DF I + + R L+ + L V+VW F +FE + G
Sbjct: 506 ISQPRLDMPEVLWKSYIDFEIEQEETERTRNLYRQLLQRT---QHVKVWISFAQFELSSG 562
Query: 492 DLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNLSRQEWL--- 548
S+ K Q +EA + + ++ R ++ W ++ +S +E +
Sbjct: 563 KEGSVAKCRQIYEEA------NKTMRNCEEKEERLMLLESWRSFEDEFGTVSDKERVDKL 616
Query: 549 ----VKNTKKVE 556
VK +KV+
Sbjct: 617 MPEKVKKRRKVQ 628
Score = 35.0 bits (79), Expect = 0.73
Identities = 28/119 (23%), Positives = 56/119 (46%), Gaps = 3/119 (2%)
Query: 392 RKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFL 451
RK F D + ++ YA L K+ + A +++E L L+YA+
Sbjct: 67 RKTFEDNIRKNRTVISNWIKYAQWEESL-KEIQRARSIYERALDVDYRNITLWLKYAEME 125
Query: 452 IRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
++ + R +++RA+++LP + + W ++ E+ G++A +V +R E EE
Sbjct: 126 MKNRQVNHARNIWDRAITTLPRVN--QFWYKYTYMEEMLGNVAGARQVFERWMEWQPEE 182
>CRN1_HUMAN (Q9BZJ0) Crooked neck-like protein 1 (Crooked neck
homolog) (hCrn) (CGI-201) (MSTP021)
Length = 848
Score = 58.9 bits (141), Expect = 5e-08
Identities = 53/229 (23%), Positives = 101/229 (43%), Gaps = 14/229 (6%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRG 351
YE+ L Y +W YA K ++ A ++ R++ LP Y Y +EE G
Sbjct: 265 YERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYTYMEEMLG 324
Query: 352 AIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVA 411
+ A++++E + A +I F R + V+ AR + + ++
Sbjct: 325 NVAGARQVFERWM--EWQPEEQAWHSYINFELRYKEVDRARTIY-ERFVLVHPDVKNWIK 381
Query: 412 YASVAFCLDKDPKMAH--NVFEAGLKHF----MHEPVYILEYADFLIRLNDDQNIRALFE 465
YA +K AH V+E ++ F M E +Y+ +A F + + +R +++
Sbjct: 382 YARFE---EKHAYFAHARKVYERAVEFFGDEHMDEHLYV-AFAKFEENQKEFERVRVIYK 437
Query: 466 RALSSLPLEDSVEVWKRFVKFEQTYGDLASMLK-VEQRRKEAFGEEATA 513
AL + +D+ E++K + FE+ +GD + + +R+ + EE A
Sbjct: 438 YALDRISKQDAQELFKNYTIFEKKFGDRRGIEDIIVSKRRFQYEEEVKA 486
Score = 51.6 bits (122), Expect = 8e-06
Identities = 103/501 (20%), Positives = 188/501 (36%), Gaps = 71/501 (14%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
+ A I+++ + P +FW +Y + N +Q+F R + + W YI
Sbjct: 292 VNHARNIWDRAITTLPRVNQFWYKYTYMEEMLGNVAGARQVFERWMEWQPEEQAWHSYIN 351
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
F + + A E F+L V D+ + W++Y F EE H
Sbjct: 352 FELRYKEVDRARTIYE-----RFVL--VHPDVKN---WIKYARF----------EEKHAY 391
Query: 143 TV-VRKVYQRAI--ITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRE 199
RKVY+RA+ H E L+ + FE E Q ++ R +Y+
Sbjct: 392 FAHARKVYERAVEFFGDEHMDEHLYVAFAKFE-------------ENQKEFERVRVIYKY 438
Query: 200 RKKFFDEIDWNMLAVPPT------GSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPK 253
+ D L T G + + + + K + ++ N +++
Sbjct: 439 ALDRISKQDAQELFKNYTIFEKKFGDRRGIEDIIVSKRRFQ----YEEEVKANPHNYDAW 494
Query: 254 SLCLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWH 313
L L+ + + E + A +N I Y+Y +W +YA +
Sbjct: 495 FDYLRLVESDAEAEAVREV------YERAIANVPPIQEKRHWKRYIY----LWINYALYE 544
Query: 314 A-KAGSIDAAIKVFQRSLKALPDSEM----LRYAYAELEESRGAIQAAKKIYENLLGDS- 367
+A + +V+Q SL+ +P + + YA+ E + + A++ +G
Sbjct: 545 ELEAKDPERTRQVYQASLELIPHKKFTFAKMWILYAQFEIRQKNLSLARRALGTSIGKCP 604
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+N +I+ LR + + FL+ +CT ++ +A + L D A
Sbjct: 605 KNKLFKVYIELELQLREFDRCRKLYEKFLEFGPE-NCTS--WIKFAELETILG-DIDRAR 660
Query: 428 NVFEAGLKHFMHEPVYIL--EYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVK 485
++E + + +L Y DF I + + R L+ R L V+VW F +
Sbjct: 661 AIYELAISQPRLDMPEVLWKSYIDFEIEQEETERTRNLYRRLLQRT---QHVKVWISFAQ 717
Query: 486 FEQTYGDLASMLKVEQRRKEA 506
FE + G S+ K Q +EA
Sbjct: 718 FELSSGKEGSLTKCRQIYEEA 738
Score = 51.6 bits (122), Expect = 8e-06
Identities = 102/489 (20%), Positives = 177/489 (35%), Gaps = 84/489 (17%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQV-PLWRCYI 81
I A IYE+ L + W +Y E M + + I+ R + +V W Y
Sbjct: 258 IQRARSIYERALDVDYRNITLWLKYAEMEMKNRQVNHARNIWDRAITTLPRVNQFWYKYT 317
Query: 82 RFIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHR 141
+ + GA ++ FE + + + A W YI F +
Sbjct: 318 YMEEMLGNVAGA------RQVFERWMEWQPEEQA----WHSYINF---------ELRYKE 358
Query: 142 MTVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERK 201
+ R +Y+R ++ H + W Y FE E + AR VY
Sbjct: 359 VDRARTIYERFVLV--HPDVKNWIKYARFE-------------EKHAYFAHARKVYERAV 403
Query: 202 KFF-----DEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVF-VNKLFWNDNHFPPKSL 255
+FF DE + A + + + KY L +S +LF N F K
Sbjct: 404 EFFGDEHMDEHLYVAFAKFEENQKEFERVRVIYKYALDRISKQDAQELFKNYTIFEKK-- 461
Query: 256 CLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAK 315
R ++ DI S +R F YE+ + H D W+DY
Sbjct: 462 ----FGDRRGIE------------DIIVSKRR--FQYEEEVKANPHNYDAWFDYLRLVES 503
Query: 316 AGSIDAAIKVFQRSLKALPDSEMLRY------------AYAELEESRGAIQAAKKIYE-- 361
+A +V++R++ +P + R+ Y ELE + +++Y+
Sbjct: 504 DAEAEAVREVYERAIANVPPIQEKRHWKRYIYLWINYALYEELEAKDP--ERTRQVYQAS 561
Query: 362 -NLLGDSENATALAHIQFIRFLRRTEGVEPARKYF-LDARKSP-SCTYHVYVAYASVAFC 418
L+ + A I + +F R + + AR+ K P + + VY+
Sbjct: 562 LELIPHKKFTFAKMWILYAQFEIRQKNLSLARRALGTSIGKCPKNKLFKVYIELELQLRE 621
Query: 419 LDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVE 478
D+ K+ E G ++ +++A+ L D RA++E A+S L+
Sbjct: 622 FDRCRKLYEKFLEFGPENCTSW----IKFAELETILGDIDRARAIYELAISQPRLDMPEV 677
Query: 479 VWKRFVKFE 487
+WK ++ FE
Sbjct: 678 LWKSYIDFE 686
Score = 35.0 bits (79), Expect = 0.73
Identities = 28/119 (23%), Positives = 56/119 (46%), Gaps = 3/119 (2%)
Query: 392 RKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFL 451
RK F D + ++ YA L K+ + A +++E L L+YA+
Sbjct: 228 RKTFEDNIRKNRTVISNWIKYAQWEESL-KEIQRARSIYERALDVDYRNITLWLKYAEME 286
Query: 452 IRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
++ + R +++RA+++LP + + W ++ E+ G++A +V +R E EE
Sbjct: 287 MKNRQVNHARNIWDRAITTLPRVN--QFWYKYTYMEEMLGNVAGARQVFERWMEWQPEE 343
>CWF4_SCHPO (P87312) Cell cycle control protein cwf4
Length = 674
Score = 58.2 bits (139), Expect = 8e-08
Identities = 52/208 (25%), Positives = 91/208 (43%), Gaps = 15/208 (7%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRG 351
+E+ L Y +W Y K +I+ A +F R++ LP + L Y Y +EE G
Sbjct: 93 FERALDVDSTYIPLWLKYIECEMKNRNINHARNLFDRAVTQLPRVDKLWYKYVYMEEMLG 152
Query: 352 AIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVA 411
I ++++E L + + +IR RR E AR + H V
Sbjct: 153 NITGCRQVFERWLKWEPDENCW--MSYIRMERRYHENERARGIY-----ERFVVVHPEVT 205
Query: 412 YASVAFCLDKDPKMAHNVFEAGL-------KHFMHEPVYILEYADFLIRLNDDQNIRALF 464
+++ A NV + L + F++E +I +A F IR + + R +F
Sbjct: 206 NWLRWARFEEECGNAANVRQVYLAAIDALGQEFLNERFFIA-FAKFEIRQKEYERARTIF 264
Query: 465 ERALSSLPLEDSVEVWKRFVKFEQTYGD 492
+ A+ +P S+E++K + FE+ +GD
Sbjct: 265 KYAIDFMPRSKSMELYKEYTHFEKQFGD 292
Score = 50.1 bits (118), Expect = 2e-05
Identities = 115/542 (21%), Positives = 199/542 (36%), Gaps = 91/542 (16%)
Query: 22 SIAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYI 81
+I A ++++ + P K W +YV + N +Q+F R L W YI
Sbjct: 119 NINHARNLFDRAVTQLPRVDKLWYKYVYMEEMLGNITGCRQVFERWLKWEPDENCWMSYI 178
Query: 82 RFIRKVNDKKGAEG---------------------QEETKKAFEFMLSYVGSDIASGPVW 120
R R+ ++ + A G +EE A Y+ + A G +
Sbjct: 179 RMERRYHENERARGIYERFVVVHPEVTNWLRWARFEEECGNAANVRQVYLAAIDALGQEF 238
Query: 121 MEYIAFLKSLPAAHPQEETHRMTVVRKVYQRAI-ITPTHHIEQLWKDYDSFESSVSQKLA 179
+ F+ Q+E R R +++ AI P +L+K+Y FE L
Sbjct: 239 LNERFFIAFAKFEIRQKEYER---ARTIFKYAIDFMPRSKSMELYKEYTHFEKQFGDHL- 294
Query: 180 KGLISEYQPKYNSARAVYRERKKF--FDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLS 237
G+ S K R Y + K +D W L + + + ++ +
Sbjct: 295 -GVESTVLDK---RRLQYEKLLKDSPYDYDTWLDLLKLEESAGDINTIRETYEKAIAKVP 350
Query: 238 VFVNKLFWNDNHFPPKSLCLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLM 297
V K W + + CLF +DV K +D A Y++ L
Sbjct: 351 EVVEKNAWRRYVYIWLNYCLF-----EEIDV--------KDVDRARK------VYQEALK 391
Query: 298 YLYH----YPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAI 353
+ H + +W YA + + ID A K R+L P ++ R Y E E++
Sbjct: 392 LIPHKKFTFAKLWLMYAMFELRQRKIDVARKTLGRALGMCPKPKLFR-GYIEFEDAIKQF 450
Query: 354 QAAKKIYEN-LLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSP--SCTYHVYV 410
+ +YE +L D E + + + + AR + A P V+
Sbjct: 451 DRCRILYEKWILYDPE--ACAPWLGYAALETKLGDSDRARALYNLAVNQPILETPELVWK 508
Query: 411 AYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADF-LIRLNDDQ----------- 458
AY F + + A ++++ L+ H V+I +A+F + L DD
Sbjct: 509 AYIDFEF-EEMEYGKARSIYQQLLRTAPHVKVWI-SFANFEIAHLEDDDEEPPNEEVASP 566
Query: 459 ----NIRALFERALSSLPLED-------SVEVWKRFVKF---EQTYGDLASMLK--VEQR 502
R +FE AL+ L + +E WK+F E T ++S++ V++R
Sbjct: 567 TAVVRARNVFENALAHLRQQGLKEERVVLLEAWKQFEAMHGTEDTRKHVSSLMPQVVKKR 626
Query: 503 RK 504
R+
Sbjct: 627 RR 628
Score = 48.1 bits (113), Expect = 8e-05
Identities = 93/488 (19%), Positives = 175/488 (35%), Gaps = 84/488 (17%)
Query: 24 AEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVP-LWRCYIR 82
A A ++E+ L + T W +Y+E M N + + +F R + +V LW Y+
Sbjct: 87 ARARSVFERALDVDSTYIPLWLKYIECEMKNRNINHARNLFDRAVTQLPRVDKLWYKYVY 146
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
+ + G ++ FE L + + WM YI + H
Sbjct: 147 MEEMLGNITGC------RQVFERWLKWEPDENC----WMSYIRM---------ERRYHEN 187
Query: 143 TVVRKVYQRAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKK 202
R +Y+R ++ H W + FE A + Y ++ + +
Sbjct: 188 ERARGIYERFVVV--HPEVTNWLRWARFEEECGN--AANVRQVYLAAIDALGQEFLNERF 243
Query: 203 FFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTG 262
F +KF K + ++F + F P+S
Sbjct: 244 FI----------------AFAKFEIRQKEYERARTIFKYAI-----DFMPRS-------- 274
Query: 263 RNAVDVMEETSIF*KR------IDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKA 316
++++ +E + F K+ ++ +KR + YE+ L + D W D A
Sbjct: 275 -KSMELYKEYTHFEKQFGDHLGVESTVLDKRRL-QYEKLLKDSPYDYDTWLDLLKLEESA 332
Query: 317 GSIDAAIKVFQRSLKALPD----SEMLRYAYAEL------EESRGAIQAAKKIYEN---L 363
G I+ + +++++ +P+ + RY Y L E + A+K+Y+ L
Sbjct: 333 GDINTIRETYEKAIAKVPEVVEKNAWRRYVYIWLNYCLFEEIDVKDVDRARKVYQEALKL 392
Query: 364 LGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCT----YHVYVAYASVAFCL 419
+ + A + + F R ++ ARK R C + Y+ +
Sbjct: 393 IPHKKFTFAKLWLMYAMFELRQRKIDVARKTL--GRALGMCPKPKLFRGYIEFEDAIKQF 450
Query: 420 DKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEV 479
D+ ++E + + L YA +L D RAL+ A++ LE V
Sbjct: 451 DR----CRILYEKWILYDPEACAPWLGYAALETKLGDSDRARALYNLAVNQPILETPELV 506
Query: 480 WKRFVKFE 487
WK ++ FE
Sbjct: 507 WKAYIDFE 514
Score = 34.7 bits (78), Expect = 0.96
Identities = 39/172 (22%), Positives = 73/172 (41%), Gaps = 8/172 (4%)
Query: 339 LRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDA 398
+RY EL++ A A+ ++E L D ++ +++I + + AR F A
Sbjct: 74 MRYGQWELDQKEFA--RARSVFERAL-DVDSTYIPLWLKYIECEMKNRNINHARNLFDRA 130
Query: 399 RKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQ 458
++ Y + L VFE LK E + + Y R ++++
Sbjct: 131 VTQLPRVDKLWYKYVYMEEMLGNITG-CRQVFERWLKWEPDENCW-MSYIRMERRYHENE 188
Query: 459 NIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
R ++ER + P V W R+ +FE+ G+ A++ +V +A G+E
Sbjct: 189 RARGIYERFVVVHP---EVTNWLRWARFEEECGNAANVRQVYLAAIDALGQE 237
>RRP5_HUMAN (Q14690) RRP5 protein homolog (Programmed cell death
protein 11)
Length = 1871
Score = 52.0 bits (123), Expect = 6e-06
Identities = 57/238 (23%), Positives = 96/238 (39%), Gaps = 41/238 (17%)
Query: 305 VWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLL 364
+W Y +H +A I+ A V +R+LK + E E+ + + A EN+
Sbjct: 1621 LWLQYMAFHLQATEIEKARAVAERALKTISFRE---------EQEKLNVWVALLNLENMY 1671
Query: 365 GDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPK 424
G E+ T + F R + EP + + H+ YA + +
Sbjct: 1672 GSQESLTKV-------FERAVQYNEPLKVFL-----------HLADIYAK-----SEKFQ 1708
Query: 425 MAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFV 484
A ++ LK F E ++Y FL+R + + +RAL LP ++ V+V +F
Sbjct: 1709 EAGELYNRMLKRFRQEKAVWIKYGAFLLRRSQAAASHRVLQRALECLPSKEHVDVIAKFA 1768
Query: 485 KFEQTYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDLDNL 542
+ E GD +R K F E T ++ DV S Y M + S D+ ++
Sbjct: 1769 QLEFQLGD-------AERAKAIF--ENTLSTYPKRTDVWSVYIDMTIKHGSQKDVRDI 1817
Score = 37.4 bits (85), Expect = 0.15
Identities = 29/105 (27%), Positives = 46/105 (43%), Gaps = 5/105 (4%)
Query: 265 AVDVMEETSIF*KRIDIASSNKRVIFT---YEQCLMYLYHYPDVWYDYATWHAKAGSIDA 321
AV E +F DI + +++ Y + L VW Y + + A
Sbjct: 1684 AVQYNEPLKVFLHLADIYAKSEKFQEAGELYNRMLKRFRQEKAVWIKYGAFLLRRSQAAA 1743
Query: 322 AIKVFQRSLKALPDSEMLRY--AYAELEESRGAIQAAKKIYENLL 364
+ +V QR+L+ LP E + +A+LE G + AK I+EN L
Sbjct: 1744 SHRVLQRALECLPSKEHVDVIAKFAQLEFQLGDAERAKAIFENTL 1788
>RRP5_YEAST (Q05022) rRNA biogenesis protein RRP5
Length = 1729
Score = 50.1 bits (118), Expect = 2e-05
Identities = 52/217 (23%), Positives = 90/217 (40%), Gaps = 33/217 (15%)
Query: 277 KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDS 336
K IDI + + +E+ L+ + VW +Y + + I+ A ++ +R+LK +
Sbjct: 1449 KTIDINTRAPESVADFERLLIGNPNSSVVWMNYMAFQLQLSEIEKARELAERALKTINFR 1508
Query: 337 EMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFL 396
E E + I A EN G E + F R + ++
Sbjct: 1509 E---------EAEKLNIWIAMLNLENTFGTEETLEEV-------FSRACQYMD------- 1545
Query: 397 DARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYI-LEYADFLIRLN 455
+Y ++ + +K K A +F+A K F E V I + + DFLI N
Sbjct: 1546 --------SYTIHTKLLGIYEISEKFDKAA-ELFKATAKKFGGEKVSIWVSWGDFLISHN 1596
Query: 456 DDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGD 492
++Q R + AL +LP + +EV ++F + E GD
Sbjct: 1597 EEQEARTILGNALKALPKRNHIEVVRKFAQLEFAKGD 1633
>CRN_DROME (P17886) Crooked neck protein
Length = 702
Score = 48.1 bits (113), Expect = 8e-05
Identities = 41/168 (24%), Positives = 77/168 (45%), Gaps = 6/168 (3%)
Query: 343 YAELEESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSP 402
YA+ EE + IQ A+ I+E L D+E+ +++ + + V AR + A
Sbjct: 82 YAQWEEQQQEIQRARSIWERAL-DNEHRNVTLWLKYAEMEMKNKQVNHARNLWDRAVTIM 140
Query: 403 SCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRA 462
+ Y + L+ + A VFE ++ E + Y +F +R + R
Sbjct: 141 PRVNQFWYKYTYMEEMLE-NVAGARQVFERWMEWQPEEQAW-QTYVNFELRYKEIDRARE 198
Query: 463 LFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
++ER + P V+ W +F +FE+++G + +V +R E FG++
Sbjct: 199 IYERFVYVHP---DVKNWIKFARFEESHGFIHGSRRVFERAVEFFGDD 243
Score = 47.0 bits (110), Expect = 2e-04
Identities = 111/568 (19%), Positives = 215/568 (37%), Gaps = 75/568 (13%)
Query: 23 IAEATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIR 82
+ A ++++ + + P +FW +Y + N +Q+F R + + W+ Y+
Sbjct: 126 VNHARNLWDRAVTIMPRVNQFWYKYTYMEEMLENVAGARQVFERWMEWQPEEQAWQTYVN 185
Query: 83 FIRKVNDKKGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRM 142
F + + + ++ +E + YV D+ + W+++ F +S H
Sbjct: 186 FELRYKE------IDRAREIYERFV-YVHPDVKN---WIKFARFEESHGFIHGS------ 229
Query: 143 TVVRKVYQRAI--ITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQ----PKYNSA--- 193
R+V++RA+ + E+L+ + FE + +I +Y PK +
Sbjct: 230 ---RRVFERAVEFFGDDYIEERLFIAFARFEEGQKEHDRARIIYKYALDHLPKDRTQELF 286
Query: 194 RAVYRERKKFFDEIDWNMLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPK 253
+A + KK+ D + V SK + +Y + + N W D
Sbjct: 287 KAYTKHEKKYGDRAGIEDVIV--------SKRKY--QYEQEVAANPTNYDAWFD------ 330
Query: 254 SLCLFLLTGRNAVDVMEETSIF*KRIDIASSNKRVIFTYEQCLMYLYHYPDVWYDYATW- 312
L L+ D + ET + A SN Y+Y +W +YA +
Sbjct: 331 --YLRLIEAEGDRDQIRET------YERAISNVPPANEKNFWRRYIY----LWINYALYE 378
Query: 313 HAKAGSIDAAIKVFQRSLKALPDSEM----LRYAYAELEESRGAIQAAKKIYENLLG-DS 367
+A + ++++ L+ +P + L YA+ E +Q A+K +G
Sbjct: 379 ELEAEDAERTRQIYKTCLELIPHKQFTFSKLWLLYAQFEIRCKELQRARKALGLAIGMCP 438
Query: 368 ENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAH 427
+ +I LR E + FL+ T+ + + L D A
Sbjct: 439 RDKLFRGYIDLEIQLREFERCRMLYEKFLEFGPENCVTWMKFAELEN----LLGDTDRAR 494
Query: 428 NVFEAGLKHFMHEPVYIL--EYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVK 485
+FE ++ + +L Y DF + L + + R L+ER L V+VW F K
Sbjct: 495 AIFELAVQQPRLDMPELLWKAYIDFEVALGETELARQLYERLLER---TQHVKVWMSFAK 551
Query: 486 FEQ--TYGDLASMLKVEQRRKEAFGEEATAASESSLQDVVSRYSFMDLWPCSSNDL-DNL 542
FE ++GD ++ + E A L D SR ++ W D D+
Sbjct: 552 FEMGLSHGDSGPDAELNVQLARRIYERANEMLR-QLGDKESRVLLLEAWRDFERDASDSQ 610
Query: 543 SRQEWLVKNTKKVEKSIMLNGTTFIDKG 570
Q+ + K ++++K + +++G
Sbjct: 611 EMQKVMDKMPRRIKKRQKIVSDNGVEEG 638
Score = 46.6 bits (109), Expect = 2e-04
Identities = 47/227 (20%), Positives = 94/227 (40%), Gaps = 10/227 (4%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRG 351
+E+ L + +W YA K ++ A ++ R++ +P Y Y +EE
Sbjct: 99 WERALDNEHRNVTLWLKYAEMEMKNKQVNHARNLWDRAVTIMPRVNQFWYKYTYMEEMLE 158
Query: 352 AIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARK-YFLDARKSPSCTYHVYV 410
+ A++++E + A ++ F R + ++ AR+ Y P +
Sbjct: 159 NVAGARQVFERWMEWQPEEQAWQ--TYVNFELRYKEIDRAREIYERFVYVHPDVKNWIKF 216
Query: 411 AYASVAFCLDKDPKMAHNVFEAGLKHFMHEPV---YILEYADFLIRLNDDQNIRALFERA 467
A + + VFE ++ F + + + +A F + R +++ A
Sbjct: 217 ARFEESHGFIHG---SRRVFERAVEFFGDDYIEERLFIAFARFEEGQKEHDRARIIYKYA 273
Query: 468 LSSLPLEDSVEVWKRFVKFEQTYGDLASMLKV-EQRRKEAFGEEATA 513
L LP + + E++K + K E+ YGD A + V +RK + +E A
Sbjct: 274 LDHLPKDRTQELFKAYTKHEKKYGDRAGIEDVIVSKRKYQYEQEVAA 320
Score = 42.0 bits (97), Expect = 0.006
Identities = 46/193 (23%), Positives = 78/193 (39%), Gaps = 18/193 (9%)
Query: 306 WYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYE---N 362
W Y + + ID A ++++R + PD + +A EES G I +++++E
Sbjct: 180 WQTYVNFELRYKEIDRAREIYERFVYVHPDVKNW-IKFARFEESHGFIHGSRRVFERAVE 238
Query: 363 LLGDSENATALAHIQFIRFLRRTEGVEPAR---KYFLDARKSPSCTYHVYVAYASVAFCL 419
GD L I F RF + + AR KY LD T ++ AY
Sbjct: 239 FFGDDYIEERL-FIAFARFEEGQKEHDRARIIYKYALD-HLPKDRTQELFKAYTKHEKKY 296
Query: 420 DKDPKMAHNVFEAGLKHFMHEPVYI--------LEYADFLIRLNDDQNIRALFERALSSL 471
D +V + K+ + V +Y + D IR +ERA+S++
Sbjct: 297 G-DRAGIEDVIVSKRKYQYEQEVAANPTNYDAWFDYLRLIEAEGDRDQIRETYERAISNV 355
Query: 472 PLEDSVEVWKRFV 484
P + W+R++
Sbjct: 356 PPANEKNFWRRYI 368
Score = 37.0 bits (84), Expect = 0.19
Identities = 48/240 (20%), Positives = 99/240 (41%), Gaps = 19/240 (7%)
Query: 280 DIASSNKRVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSE-- 337
D+ S ++ + YEQ + D W+DY G D + ++R++ +P +
Sbjct: 304 DVIVSKRK--YQYEQEVAANPTNYDAWFDYLRLIEAEGDRDQIRETYERAISNVPPANEK 361
Query: 338 ---------MLRYAYAELEESRGAIQAAKKIYEN---LLGDSENATALAHIQFIRFLRRT 385
+ YA E E+ A + ++IY+ L+ + + + + +F R
Sbjct: 362 NFWRRYIYLWINYALYEELEAEDA-ERTRQIYKTCLELIPHKQFTFSKLWLLYAQFEIRC 420
Query: 386 EGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYIL 445
+ ++ ARK L ++ Y + L ++ + ++E L+ V +
Sbjct: 421 KELQRARKA-LGLAIGMCPRDKLFRGYIDLEIQL-REFERCRMLYEKFLEFGPENCVTWM 478
Query: 446 EYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKE 505
++A+ L D RA+FE A+ L+ +WK ++ FE G+ ++ +R E
Sbjct: 479 KFAELENLLGDTDRARAIFELAVQQPRLDMPELLWKAYIDFEVALGETELARQLYERLLE 538
>XAB2_RAT (Q99PK0) XPA-binding protein 2 (Adapter protein ATH-55)
Length = 855
Score = 44.3 bits (103), Expect = 0.001
Identities = 44/232 (18%), Positives = 94/232 (39%), Gaps = 23/232 (9%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY--AYAELEES 349
+E+ ++++ P +W DY + G + + F R+L+ALP ++ R Y S
Sbjct: 99 HERAFVFMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFLRS 158
Query: 350 RGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFL----DARKSPSCT 405
+ A + Y L S + ++I +L+ ++ ++ A + D R
Sbjct: 159 HPLPETAVRGYRRFLKLSPESAE----EYIEYLKSSDRLDEAAQRLATVVNDERFVSKAG 214
Query: 406 YHVYVAYASVAFCLDKDPKMAHN-----VFEAGLKHFMHEPVYI-LEYADFLIRLNDDQN 459
Y + + + ++P + + GL F + + AD+ IR +
Sbjct: 215 KSNYQLWHELCDLISQNPDKVQSLNVDAIIRGGLTRFTDQLGKLWCSLADYYIRSGHFEK 274
Query: 460 IRALFERALSS-LPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
R ++E A+ + + + D +V+ + +FE+ SM+ + G E
Sbjct: 275 ARDVYEEAIRTVMTVRDFTQVFDSYAQFEE------SMIAAKMETASELGRE 320
Score = 37.0 bits (84), Expect = 0.19
Identities = 44/233 (18%), Positives = 93/233 (39%), Gaps = 43/233 (18%)
Query: 299 LYHYP---DVWYDYAT-WHAKAGS--IDAAIKVFQRSLKALPD--SEMLRYAYAELEESR 350
L+ +P D+W Y T + ++ G ++ A +F+++L P ++ L YA+LEE
Sbjct: 547 LFKWPNVSDIWSTYLTKFISRYGGRKLERARDLFEQALDGCPPKYAKTLYLLYAQLEEEW 606
Query: 351 GAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYV 410
G + A +Y+ R T VEPA++Y + +++Y+
Sbjct: 607 GLARHAMAVYD---------------------RATRAVEPAQQYDM---------FNIYI 636
Query: 411 AYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS 470
A+ + + + E + H L +AD +L + RA++
Sbjct: 637 KRAAEIYGVTHTRGIYQKAIE--VLSDEHAREMCLRFADMECKLGEIDRARAIYSFCSQI 694
Query: 471 LPLEDSVEVWKRFVKFEQTYGD---LASMLKVEQRRKEAFGEEATAASESSLQ 520
+ W+ + FE +G+ + ML++ + + + + + L+
Sbjct: 695 CDPRTTGAFWQTWKDFEVRHGNEDTIREMLRIRRSVQATYNTQVNFMASQMLK 747
Score = 36.6 bits (83), Expect = 0.25
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 11/123 (8%)
Query: 378 FIRFLRRTEGVEPAR--KYFLDARKSPSCTYHVYVAY-----ASVAFCLDKDPKM--AHN 428
++R++ +G R + + A K C+Y ++ Y A V DP +N
Sbjct: 38 WLRYIEFKQGAPKPRLNQLYERALKLLPCSYKLWYRYLKARRAQVKHRCVTDPAYEDVNN 97
Query: 429 VFEAGLKHFMHE-PVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFE 487
E FMH+ P L+Y FL+ + R F+RAL +LP+ +W +++F
Sbjct: 98 CHERAFV-FMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFL 156
Query: 488 QTY 490
+++
Sbjct: 157 RSH 159
Score = 32.0 bits (71), Expect = 6.2
Identities = 49/192 (25%), Positives = 77/192 (39%), Gaps = 35/192 (18%)
Query: 315 KAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALA 374
+A D + V R K+L ML A+LEES G Q+ K +Y+ +L D AT
Sbjct: 466 RAEYFDGSEPVQNRVYKSLKVWSML----ADLEESLGTFQSTKAVYDRIL-DLRIATPQI 520
Query: 375 HIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGL 434
I + FL KYF ++ K AY L K P ++ +++ L
Sbjct: 521 VINYAMFLEE-------HKYFEESFK----------AYER-GISLFKWPNVS-DIWSTYL 561
Query: 435 KHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLA 494
F I Y + + R LFE+AL P + + ++ + + E+ +G
Sbjct: 562 TKF------ISRYGGRKL-----ERARDLFEQALDGCPPKYAKTLYLLYAQLEEEWGLAR 610
Query: 495 SMLKVEQRRKEA 506
+ V R A
Sbjct: 611 HAMAVYDRATRA 622
>XAB2_MOUSE (Q9DCD2) XPA-binding protein 2
Length = 855
Score = 44.3 bits (103), Expect = 0.001
Identities = 44/232 (18%), Positives = 94/232 (39%), Gaps = 23/232 (9%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY--AYAELEES 349
+E+ ++++ P +W DY + G + + F R+L+ALP ++ R Y S
Sbjct: 99 HERAFVFMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFLRS 158
Query: 350 RGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFL----DARKSPSCT 405
+ A + Y L S + ++I +L+ ++ ++ A + D R
Sbjct: 159 HPLPETAVRGYRRFLKLSPESAE----EYIEYLKSSDRLDEAAQRLATVVNDERFVSKAG 214
Query: 406 YHVYVAYASVAFCLDKDPKMAHN-----VFEAGLKHFMHEPVYI-LEYADFLIRLNDDQN 459
Y + + + ++P + + GL F + + AD+ IR +
Sbjct: 215 KSNYQLWHELCDLISQNPDKVQSLNVDAIIRGGLTRFTDQLGKLWCSLADYYIRSGHFEK 274
Query: 460 IRALFERALSS-LPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
R ++E A+ + + + D +V+ + +FE+ SM+ + G E
Sbjct: 275 ARDVYEEAIRTVMTVRDFTQVFDSYAQFEE------SMIAAKMETASELGRE 320
Score = 37.0 bits (84), Expect = 0.19
Identities = 44/233 (18%), Positives = 93/233 (39%), Gaps = 43/233 (18%)
Query: 299 LYHYP---DVWYDYAT-WHAKAGS--IDAAIKVFQRSLKALPD--SEMLRYAYAELEESR 350
L+ +P D+W Y T + ++ G ++ A +F+++L P ++ L YA+LEE
Sbjct: 547 LFKWPNVSDIWSTYLTKFISRYGGRKLERARDLFEQALDGCPPKYAKTLYLLYAQLEEEW 606
Query: 351 GAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYV 410
G + A +Y+ R T VEPA++Y + +++Y+
Sbjct: 607 GLARHAMAVYD---------------------RATRAVEPAQQYDM---------FNIYI 636
Query: 411 AYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS 470
A+ + + + E + H L +AD +L + RA++
Sbjct: 637 KRAAEIYGVTHTRGIYQKAIE--VLSDEHAREMCLRFADMECKLGEIDRARAIYSFCSQI 694
Query: 471 LPLEDSVEVWKRFVKFEQTYGD---LASMLKVEQRRKEAFGEEATAASESSLQ 520
+ W+ + FE +G+ + ML++ + + + + + L+
Sbjct: 695 CDPRTTGAFWQTWKDFEVRHGNEDTIREMLRIRRSVQATYNTQVNFMASQMLK 747
Score = 36.6 bits (83), Expect = 0.25
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 11/123 (8%)
Query: 378 FIRFLRRTEGVEPAR--KYFLDARKSPSCTYHVYVAY-----ASVAFCLDKDPKM--AHN 428
++R++ +G R + + A K C+Y ++ Y A V DP +N
Sbjct: 38 WLRYIEFKQGAPKPRLNQLYERALKLLPCSYKLWYRYLKARRAQVKHRCVTDPAYEDVNN 97
Query: 429 VFEAGLKHFMHE-PVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFE 487
E FMH+ P L+Y FL+ + R F+RAL +LP+ +W +++F
Sbjct: 98 CHERAFV-FMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFL 156
Query: 488 QTY 490
+++
Sbjct: 157 RSH 159
Score = 32.0 bits (71), Expect = 6.2
Identities = 49/192 (25%), Positives = 77/192 (39%), Gaps = 35/192 (18%)
Query: 315 KAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALA 374
+A D + V R K+L ML A+LEES G Q+ K +Y+ +L D AT
Sbjct: 466 RAEYFDGSEPVQNRVYKSLKVWSML----ADLEESLGTFQSTKAVYDRIL-DLRIATPQI 520
Query: 375 HIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGL 434
I + FL KYF ++ K AY L K P ++ +++ L
Sbjct: 521 VINYAMFLEE-------HKYFEESFK----------AYER-GISLFKWPNVS-DIWSTYL 561
Query: 435 KHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLA 494
F I Y + + R LFE+AL P + + ++ + + E+ +G
Sbjct: 562 TKF------ISRYGGRKL-----ERARDLFEQALDGCPPKYAKTLYLLYAQLEEEWGLAR 610
Query: 495 SMLKVEQRRKEA 506
+ V R A
Sbjct: 611 HAMAVYDRATRA 622
>XAB2_HUMAN (Q9HCS7) XPA-binding protein 2 (HCNP protein) (PP3898)
Length = 855
Score = 44.3 bits (103), Expect = 0.001
Identities = 44/232 (18%), Positives = 94/232 (39%), Gaps = 23/232 (9%)
Query: 292 YEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRY--AYAELEES 349
+E+ ++++ P +W DY + G + + F R+L+ALP ++ R Y S
Sbjct: 99 HERAFVFMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFLRS 158
Query: 350 RGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFL----DARKSPSCT 405
+ A + Y L S + ++I +L+ ++ ++ A + D R
Sbjct: 159 HPLPETAVRGYRRFLKLSPESAE----EYIEYLKSSDRLDEAAQRLATVVNDERFVSKAG 214
Query: 406 YHVYVAYASVAFCLDKDPKMAHN-----VFEAGLKHFMHEPVYI-LEYADFLIRLNDDQN 459
Y + + + ++P + + GL F + + AD+ IR +
Sbjct: 215 KSNYQLWHELCDLISQNPDKVQSLNVDAIIRGGLTRFTDQLGKLWCSLADYYIRSGHFEK 274
Query: 460 IRALFERALSS-LPLEDSVEVWKRFVKFEQTYGDLASMLKVEQRRKEAFGEE 510
R ++E A+ + + + D +V+ + +FE+ SM+ + G E
Sbjct: 275 ARDVYEEAIRTVMTVRDFTQVFDSYAQFEE------SMIAAKMETASELGRE 320
Score = 39.3 bits (90), Expect = 0.039
Identities = 46/233 (19%), Positives = 93/233 (39%), Gaps = 43/233 (18%)
Query: 299 LYHYP---DVWYDYAT-WHAKAGS--IDAAIKVFQRSLKALPD--SEMLRYAYAELEESR 350
L+ +P D+W Y T + A+ G ++ A +F+++L P ++ L YA+LEE
Sbjct: 547 LFKWPNVSDIWSTYLTKFIARYGGRKLERARDLFEQALDGCPPKYAKTLYLLYAQLEEEW 606
Query: 351 GAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYV 410
G + A +YE R T VEPA++Y + +++Y+
Sbjct: 607 GLARHAMAVYE---------------------RATRAVEPAQQYDM---------FNIYI 636
Query: 411 AYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS 470
A+ + + + E + H L +AD +L + RA++
Sbjct: 637 KRAAEIYGVTHTRGIYQKAIE--VLSDEHAREMCLRFADMECKLGEIDRARAIYSFCSQI 694
Query: 471 LPLEDSVEVWKRFVKFEQTYGD---LASMLKVEQRRKEAFGEEATAASESSLQ 520
+ W+ + FE +G+ + ML++ + + + + + L+
Sbjct: 695 CDPRTTGAFWQTWKDFEVRHGNEDTIKEMLRIRRSVQATYNTQVNFMASQMLK 747
Score = 36.6 bits (83), Expect = 0.25
Identities = 31/123 (25%), Positives = 56/123 (45%), Gaps = 11/123 (8%)
Query: 378 FIRFLRRTEGVEPAR--KYFLDARKSPSCTYHVYVAY-----ASVAFCLDKDPKM--AHN 428
++R++ +G R + + A K C+Y ++ Y A V DP +N
Sbjct: 38 WLRYIEFKQGAPKPRLNQLYERALKLLPCSYKLWYRYLKARRAQVKHRCVTDPAYEDVNN 97
Query: 429 VFEAGLKHFMHE-PVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFE 487
E FMH+ P L+Y FL+ + R F+RAL +LP+ +W +++F
Sbjct: 98 CHERAFV-FMHKMPRLWLDYCQFLMDQGRVTHTRRTFDRALRALPITQHSRIWPLYLRFL 156
Query: 488 QTY 490
+++
Sbjct: 157 RSH 159
Score = 33.1 bits (74), Expect = 2.8
Identities = 49/192 (25%), Positives = 78/192 (40%), Gaps = 35/192 (18%)
Query: 315 KAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKKIYENLLGDSENATALA 374
+A D + V R K+L ML A+LEES G Q+ K +Y+ +L D AT
Sbjct: 466 RAEYFDGSEPVQNRVYKSLKVWSML----ADLEESLGTFQSTKAVYDRIL-DLRIATPQI 520
Query: 375 HIQFIRFLRRTEGVEPARKYFLDARKSPSCTYHVYVAYASVAFCLDKDPKMAHNVFEAGL 434
I + FL KYF ++ K AY L K P ++ +++ L
Sbjct: 521 VINYAMFLEE-------HKYFEESFK----------AYER-GISLFKWPNVS-DIWSTYL 561
Query: 435 KHFMHEPVYILEYADFLIRLNDDQNIRALFERALSSLPLEDSVEVWKRFVKFEQTYGDLA 494
F I Y + + R LFE+AL P + + ++ + + E+ +G
Sbjct: 562 TKF------IARYGGRKL-----ERARDLFEQALDGCPPKYAKTLYLLYAQLEEEWGLAR 610
Query: 495 SMLKVEQRRKEA 506
+ V +R A
Sbjct: 611 HAMAVYERATRA 622
>CWF3_SCHPO (Q9P7R9) Cell cycle control protein cwf3
Length = 790
Score = 38.9 bits (89), Expect = 0.051
Identities = 44/204 (21%), Positives = 86/204 (41%), Gaps = 11/204 (5%)
Query: 299 LYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEESRGAIQAAKK 358
L+ +W Y G+I+ K++ R + + + YA L E + + K
Sbjct: 476 LHKSSKIWMYYLDLEESVGTIETTRKLYDRVFELKIATPQVVVNYANLLEENAYFEDSFK 535
Query: 359 IYEN--LLGDSENATALAHIQFIRFLRRTEG--VEPARKYFLDARKS--PSCTYHVYVAY 412
IYE L A L ++ +F++R +G +E R F A + P + +Y+ Y
Sbjct: 536 IYERGVALFSYPVAFELWNLYLTKFVKRYQGTHMERTRDLFEQALEGCPPEFSKSIYLLY 595
Query: 413 ASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNI---RALFERALS 469
A K + + +A K + + I Y L+++ + + R ++E+A+
Sbjct: 596 ADFEEKFGKAKRSISILEKAADKVKTADRLAI--YNVLLVKVALNYGVLATRTVYEKAIE 653
Query: 470 SLPLEDSVEVWKRFVKFEQTYGDL 493
SL + ++ RF + E G++
Sbjct: 654 SLSDSEVKDMCLRFAEMETKLGEI 677
Score = 38.1 bits (87), Expect = 0.087
Identities = 23/83 (27%), Positives = 37/83 (43%), Gaps = 11/83 (13%)
Query: 411 AYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFERALSS 470
A+ASV C ++ + H + PV Y FL++ + IR F AL +
Sbjct: 90 AFASVNDCFERSLILLHKM-----------PVIWKLYLQFLMKQPNVTKIRCTFNSALRA 138
Query: 471 LPLEDSVEVWKRFVKFEQTYGDL 493
LP+ ++W F K+ + G L
Sbjct: 139 LPVTQHDDIWDMFTKYAEDIGGL 161
Score = 36.2 bits (82), Expect = 0.33
Identities = 47/221 (21%), Positives = 81/221 (36%), Gaps = 59/221 (26%)
Query: 305 VWYDYATWHAKAGSIDAAIKV---------------FQRSLKA---LPDSEMLRYAYAEL 346
VW D+A + + DAA K+ F SL L S + Y +L
Sbjct: 430 VWIDWAEMELRHQNFDAARKLIGDAVHAPRKSHISFFDESLSPQVRLHKSSKIWMYYLDL 489
Query: 347 EESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTY 406
EES G I+ +K+Y+ + + + AT + + L YF D+ K
Sbjct: 490 EESVGTIETTRKLYDRVF-ELKIATPQVVVNYANLLEENA-------YFEDSFK------ 535
Query: 407 HVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHE---PVYILEYADFLIRLNDD--QNIR 461
++E G+ F + ++ L F+ R + R
Sbjct: 536 ----------------------IYERGVALFSYPVAFELWNLYLTKFVKRYQGTHMERTR 573
Query: 462 ALFERALSSLPLEDSVEVWKRFVKFEQTYGDLASMLKVEQR 502
LFE+AL P E S ++ + FE+ +G + + ++
Sbjct: 574 DLFEQALEGCPPEFSKSIYLLYADFEEKFGKAKRSISILEK 614
>PR39_YEAST (P39682) Pre-mRNA processing protein PRP39
Length = 629
Score = 36.6 bits (83), Expect = 0.25
Identities = 59/318 (18%), Positives = 112/318 (34%), Gaps = 70/318 (22%)
Query: 32 QLLQLYPTAAKFWKQYVEA-HMAVNNDDAIKQIFSRCLLNCLQVPLWRCYIRFIRKVNDK 90
Q+L+ YP FWK++ + +I + + + LW Y+ + N
Sbjct: 79 QILRKYPLLFGFWKRFATIEYQLFGLKKSIAVLATSVKWFPTSLELWCDYLNVLCVNNPN 138
Query: 91 KGAEGQEETKKAFEFMLSYVGSDIASGPVWMEYIAFLKSLPAAHPQEETHRMTVVRKVYQ 150
+ + + FE +G S P W ++I F Q+ H V+++Y+
Sbjct: 139 E----TDFIRNNFEIAKDLIGKQFLSHPFWDKFIEFEVG------QKNWHN---VQRIYE 185
Query: 151 RAIITPTHHIEQLWKDYDSFESSVSQKLAKGLISEYQPKYNSARAVYRERKKFFDEIDWN 210
I P H + + Y F + + K + + V R+ + +EI W
Sbjct: 186 YIIEVPLHQYARFFTSYKKFLNEKNLKTTRNI-----------DIVLRKTQTTVNEI-W- 232
Query: 211 MLAVPPTGSHKASKFLFLCKYWLSLLSVFVNKLFWNDNHFPPKSLCLFLLTGRNAVDVME 270
F K F G+ D +E
Sbjct: 233 --------------------------------------QFESKIKQPFFNLGQVLNDDLE 254
Query: 271 ETSIF*KRIDIASSN---KRVIFTYEQCLMYLYHYPDVWYDYATWHAKAG-SIDAAIKVF 326
S + K + S + + V+ +++CL+ ++ + W Y W K S + + ++
Sbjct: 255 NWSRYLKFVTDPSKSLDKEFVMSVFDRCLIPCLYHENTWMMYIKWLTKKNISDEVVVDIY 314
Query: 327 QRSLKALP-DSEMLRYAY 343
Q++ LP D + LRY +
Sbjct: 315 QKANTFLPLDFKTLRYDF 332
>CX56_CHICK (P29415) Gap junction Cx56 protein (Connexin 56)
Length = 510
Score = 36.2 bits (82), Expect = 0.33
Identities = 51/188 (27%), Positives = 76/188 (40%), Gaps = 21/188 (11%)
Query: 571 PVASISTTSSKVVYPDTSKM--LIYDPKHNPGTGAAGTNAFDEILKATPPALVAFLANLP 628
P ++TT+ V PDT + L+ P AA + PP+ A +A+ P
Sbjct: 287 PPVVVTTTAPAPVLPDTRAVTPLLAPVTMAPYYAAAAP-------RTRPPSNTASMASYP 339
Query: 629 SVDGPTPNVDIVLSICLQSDLPTGQSVKVGIPSQLPAGPAPATSELSGSSKSHPVQSGLS 688
V P P ++ PT S V IP+ +P P PA +SKS+ + + +
Sbjct: 340 -VAPPVPENRH------RAVTPTPVSTPVTIPTPIPT-PTPAIINYF-NSKSNALAAEQN 390
Query: 689 HMQPGRKQYGKRKQLDSQEEDDTKSVQSQPLPQDAFRIRQFQKARAGSTSQTGSVSYGSA 748
+ +Q GK S T S PLP+ + Q AG T + ++
Sbjct: 391 WVNMAAEQQGKAPS--SSAGSSTPSSVRHPLPEQEEPLEQLLPLPAGPPITTTNSGSSTS 448
Query: 749 LSGDLSGS 756
LSG SGS
Sbjct: 449 LSG-ASGS 455
>PRP1_SCHPO (Q12381) Pre-mRNA splicing factor prp1
Length = 906
Score = 35.4 bits (80), Expect = 0.56
Identities = 21/81 (25%), Positives = 39/81 (47%), Gaps = 3/81 (3%)
Query: 26 ATPIYEQLLQLYPTAAKFWKQYVEAHMAVNNDDAIKQIFSRCLLNCLQVP-LWRCYIRFI 84
A ++ L++YP + K W + VE +++ I + + +C + LW Y +
Sbjct: 529 ARAVFAFSLRVYPKSEKLWLRAVELEKLYGTTESVCSILEKAVESCPKAEILWLLYAKER 588
Query: 85 RKVNDKKGAEGQEETKKAFEF 105
+ VND GA + +AFE+
Sbjct: 589 KNVNDIAGA--RNILGRAFEY 607
>PCLO_CHICK (Q9PU36) Piccolo protein (Aczonin) (Fragment)
Length = 5120
Score = 35.0 bits (79), Expect = 0.73
Identities = 22/65 (33%), Positives = 36/65 (54%), Gaps = 4/65 (6%)
Query: 656 KVGIPSQLP--AGPAPATSELSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKS 713
+ G Q+P AGP +S+ +G++KS Q GL+ +P +Q G K L ++ T+
Sbjct: 301 QTGPVKQVPPQAGPTKPSSQTAGAAKSLAQQPGLT--KPPGQQPGPEKPLQQKQASTTQP 358
Query: 714 VQSQP 718
V+S P
Sbjct: 359 VESTP 363
>TI60_HUMAN (Q92993) Histone acetyltransferase HTATIP (EC 2.3.1.48)
(60 kDa Tat interactive protein) (Tip60) (HIV-1 Tat
interactive protein) (cPLA(2) interacting protein)
Length = 513
Score = 34.3 bits (77), Expect = 1.3
Identities = 26/105 (24%), Positives = 41/105 (38%), Gaps = 7/105 (6%)
Query: 618 PALVAFLANLP----SVDGPTPNVDIVLSICLQSDLPTGQSVKVGIPSQLPAGPAPATSE 673
P + NLP ++ G P+ + S CLQ P +S K + PA P P+ +
Sbjct: 109 PVQITLRFNLPKEREAIPGGEPDQPLSSSSCLQ---PNHRSTKRKVEVVSPATPVPSETA 165
Query: 674 LSGSSKSHPVQSGLSHMQPGRKQYGKRKQLDSQEEDDTKSVQSQP 718
+ + QPGRK+ D +D + + S P
Sbjct: 166 PASVFPQNGAARRAVAAQPGRKRKSNCLGTDEDSQDSSDGIPSAP 210
>OGT_CAEEL (O18158) UDP-N-acetylglucosamine--peptide
N-acetylglucosaminyltransferase (EC 2.4.1.-) (O-GlcNAc)
(OGT)
Length = 1151
Score = 34.3 bits (77), Expect = 1.3
Identities = 24/110 (21%), Positives = 52/110 (46%), Gaps = 1/110 (0%)
Query: 289 IFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAELEE 348
I TY++ + H+PD + + A + GS+ A +++ ++L+ P + A ++
Sbjct: 383 IDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCPTHADSQNNLANIKR 442
Query: 349 SRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDA 398
+G I+ A ++Y L + A AH L++ + A ++ +A
Sbjct: 443 EQGKIEDATRLYLKAL-EIYPEFAAAHSNLASILQQQGKLNDAILHYKEA 491
>PRPU_HUMAN (O94906) U5 snRNP-associated 102 kDa protein (U5-102 kDa
protein)
Length = 941
Score = 33.9 bits (76), Expect = 1.6
Identities = 41/207 (19%), Positives = 82/207 (38%), Gaps = 7/207 (3%)
Query: 287 RVIFTYEQCLMYLYHYPDVWYDYATWHAKAGSIDAAIKVFQRSLKALPDSEMLRYAYAEL 346
R I+ Y L VW A + G+ ++ + QR++ P +E+L A+
Sbjct: 560 RAIYAY--ALQVFPSKKSVWLRAAYFEKNHGTRESLEALLQRAVAHCPKAEVLWLMGAKS 617
Query: 347 EESRGAIQAAKKIYENLLGDSENATALAHIQFIRFLRRTEGVEPARKYFLDARKSPSCTY 406
+ G + AA+ I + N+ + + ++ + E AR+ AR S + T
Sbjct: 618 KWLAGDVPAARSILALAFQANPNSEEI-WLAAVKLESENDEYERARRLLAKARSS-APTA 675
Query: 407 HVYVAYASVAFCLDKDPKMAHNVFEAGLKHFMHEPVYILEYADFLIRLNDDQNIRALFER 466
V++ + + D + + A ++ E L+H+ P + + + R + +
Sbjct: 676 RVFMKSVKLEWVQD-NIRAAQDLCEEALRHYEDFPKLWMMKGQIEEQKEMMEKAREAYNQ 734
Query: 467 ALSSLPLEDSVEVWKRFVKFEQTYGDL 493
L P S +W + E+ G L
Sbjct: 735 GLKKCP--HSTPLWLLLSRLEEKIGQL 759
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 86,240,231
Number of Sequences: 164201
Number of extensions: 3617148
Number of successful extensions: 9587
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 9415
Number of HSP's gapped (non-prelim): 135
length of query: 757
length of database: 59,974,054
effective HSP length: 118
effective length of query: 639
effective length of database: 40,598,336
effective search space: 25942336704
effective search space used: 25942336704
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC146910.2


