

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
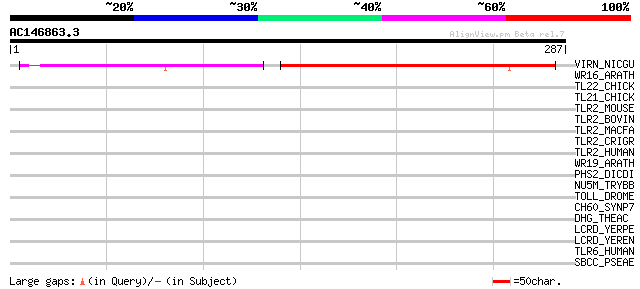
Query= AC146863.3 + phase: 0
(287 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 136 5e-32
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 42 0.002
TL22_CHICK (Q9DGB6) Toll-like receptor 2 type 2 precursor 37 0.073
TL21_CHICK (Q9DD78) Toll-like receptor 2 type 1 precursor 37 0.073
TLR2_MOUSE (Q9QUN7) Toll-like receptor 2 precursor 35 0.28
TLR2_BOVIN (Q95LA9) Toll-like receptor 2 precursor 34 0.47
TLR2_MACFA (Q95M53) Toll-like receptor 2 precursor 33 0.62
TLR2_CRIGR (Q9R1F8) Toll-like receptor 2 precursor 33 0.62
TLR2_HUMAN (O60603) Toll-like receptor 2 precursor (Toll/interle... 33 1.1
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 32 1.4
PHS2_DICDI (P34114) Glycogen phosphorylase 2 (EC 2.4.1.1) (GP2) 32 1.8
NU5M_TRYBB (P04540) NADH-ubiquinone oxidoreductase chain 5 (EC 1... 32 2.3
TOLL_DROME (P08953) Toll protein precursor 31 4.0
CH60_SYNP7 (P22879) 60 kDa chaperonin (Protein Cpn60) (groEL pro... 31 4.0
DHG_THEAC (P13203) Glucose 1-dehydrogenase (EC 1.1.1.47) 30 5.2
LCRD_YERPE (P31487) Low calcium response locus protein D 30 6.8
LCRD_YEREN (P21210) Low calcium response locus protein D 30 6.8
TLR6_HUMAN (Q9Y2C9) Toll-like receptor 6 precursor 30 8.9
SBCC_PSEAE (Q9HWB8) Nuclease sbcCD subunit C 30 8.9
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 136 bits (343), Expect = 5e-32
Identities = 66/144 (45%), Positives = 91/144 (62%), Gaps = 2/144 (1%)
Query: 141 NYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYI 200
+YDVF+SFRG DTR FT HL+ L +GI F+DD +L+ G I L +AIE SQ I
Sbjct: 11 SYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCKAIEESQFAI 70
Query: 201 VVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKH--G 258
VVFS+NYA+S WCL EL I+ C ++ + ++PIFYDVDPS V+ Q + +A +H
Sbjct: 71 VVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESFAKAFEEHETK 130
Query: 259 FKHGLNMVHRWRETLTQVGNISSS 282
+K + + RWR L + N+ S
Sbjct: 131 YKDDVEGIQRWRIALNEAANLKGS 154
Score = 82.0 bits (201), Expect = 2e-15
Identities = 50/131 (38%), Positives = 71/131 (54%), Gaps = 10/131 (7%)
Query: 6 PGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIFYDVDP 65
PG+L AIE+SQ IVVFS+N+A S +CL EL I+ C + + ++PIFYDVDP
Sbjct: 56 PGELC-----KAIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDP 110
Query: 66 SEVRKQSGGYGESL----AKLEEIAPQVQRWREALQLVGNISG-WDLCHKPQHAELENII 120
S VR Q + ++ K ++ +QRWR AL N+ G D K + I+
Sbjct: 111 SHVRNQKESFAKAFEEHETKYKDDVEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIV 170
Query: 121 EHINILGCKFS 131
+ I+ CK S
Sbjct: 171 DQISSKLCKIS 181
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 42.0 bits (97), Expect = 0.002
Identities = 29/108 (26%), Positives = 52/108 (47%), Gaps = 12/108 (11%)
Query: 152 DTRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLYIVVFSKNYASST 211
+ R++F HL ALQR+G+N D + + ++ +E +++ +++ N T
Sbjct: 15 EVRYSFVSHLSKALQRKGVN----DVFIDSDDSLSNESQSMVERARVSVMILPGN---RT 67
Query: 212 WCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGYGEALSKHGF 259
L +L +L C K + ++P+ Y V SE + S AL GF
Sbjct: 68 VSLDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLS-----ALDSKGF 110
>TL22_CHICK (Q9DGB6) Toll-like receptor 2 type 2 precursor
Length = 781
Score = 36.6 bits (83), Expect = 0.073
Identities = 22/84 (26%), Positives = 40/84 (47%), Gaps = 5/84 (5%)
Query: 139 EINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFR---DDTKLKKGEFIAPGLFRAIEA 195
+I YD FVS+ D+ N+ +++ + FR G++I + +IE
Sbjct: 635 DICYDAFVSYSENDS--NWVENIMVQQLEQACPPFRLCLHKRDFVPGKWIVDNIIDSIEK 692
Query: 196 SQLYIVVFSKNYASSTWCLRELEY 219
S + V S+++ S WC EL++
Sbjct: 693 SHKTLFVLSEHFVQSEWCKYELDF 716
>TL21_CHICK (Q9DD78) Toll-like receptor 2 type 1 precursor
Length = 793
Score = 36.6 bits (83), Expect = 0.073
Identities = 22/84 (26%), Positives = 40/84 (47%), Gaps = 5/84 (5%)
Query: 139 EINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFR---DDTKLKKGEFIAPGLFRAIEA 195
+I YD FVS+ D+ N+ +++ + FR G++I + +IE
Sbjct: 647 DICYDAFVSYSENDS--NWVENIMVQQLEQACPPFRLCLHKRDFVPGKWIVDNIIDSIEK 704
Query: 196 SQLYIVVFSKNYASSTWCLRELEY 219
S + V S+++ S WC EL++
Sbjct: 705 SHKTLFVLSEHFVQSEWCKYELDF 728
>TLR2_MOUSE (Q9QUN7) Toll-like receptor 2 precursor
Length = 784
Score = 34.7 bits (78), Expect = 0.28
Identities = 21/84 (25%), Positives = 39/84 (46%), Gaps = 5/84 (5%)
Query: 139 EINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFR---DDTKLKKGEFIAPGLFRAIEA 195
++ YD FVS+ D+ ++ ++L F+ G++I + +IE
Sbjct: 638 DVCYDAFVSYSEQDS--HWVENLMVQQLENSDPPFKLCLHKRDFVPGKWIIDNIIDSIEK 695
Query: 196 SQLYIVVFSKNYASSTWCLRELEY 219
S + V S+N+ S WC EL++
Sbjct: 696 SHKTVFVLSENFVRSEWCKYELDF 719
Score = 29.6 bits (65), Expect = 8.9
Identities = 15/42 (35%), Positives = 22/42 (51%)
Query: 3 DYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAY 44
D+ PG +I +IE S + V S+NF S +C EL +
Sbjct: 678 DFVPGKWIIDNIIDSIEKSHKTVFVLSENFVRSEWCKYELDF 719
>TLR2_BOVIN (Q95LA9) Toll-like receptor 2 precursor
Length = 784
Score = 33.9 bits (76), Expect = 0.47
Identities = 25/88 (28%), Positives = 39/88 (43%), Gaps = 13/88 (14%)
Query: 139 EINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFRDDTKLK-------KGEFIAPGLFR 191
+I YD FVS+ D+ + L + + F KL G++I +
Sbjct: 638 DICYDAFVSYSERDS------YWVENLMVQELEHFNPPFKLCLHKRDFIPGKWIIDNIID 691
Query: 192 AIEASQLYIVVFSKNYASSTWCLRELEY 219
+IE S I V S+N+ S WC EL++
Sbjct: 692 SIEKSHKTIFVLSENFVKSEWCKYELDF 719
Score = 30.4 bits (67), Expect = 5.2
Identities = 16/42 (38%), Positives = 22/42 (52%)
Query: 3 DYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAY 44
D+ PG +I +IE S I V S+NF S +C EL +
Sbjct: 678 DFIPGKWIIDNIIDSIEKSHKTIFVLSENFVKSEWCKYELDF 719
>TLR2_MACFA (Q95M53) Toll-like receptor 2 precursor
Length = 784
Score = 33.5 bits (75), Expect = 0.62
Identities = 24/91 (26%), Positives = 37/91 (40%), Gaps = 19/91 (20%)
Query: 139 EINYDVFVSFRGPDTRF----------NFTDHLFAALQRRGINAFRDDTKLKKGEFIAPG 188
+I YD FVS+ D + NF L +R G++I
Sbjct: 638 DICYDAFVSYSERDAYWVENLMVQELENFNPPFKLCLHKRDFIP---------GKWIIDN 688
Query: 189 LFRAIEASQLYIVVFSKNYASSTWCLRELEY 219
+ +IE S + V S+N+ S WC EL++
Sbjct: 689 IIDSIEKSHKTVFVLSENFVKSEWCKYELDF 719
Score = 30.0 bits (66), Expect = 6.8
Identities = 15/42 (35%), Positives = 22/42 (51%)
Query: 3 DYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAY 44
D+ PG +I +IE S + V S+NF S +C EL +
Sbjct: 678 DFIPGKWIIDNIIDSIEKSHKTVFVLSENFVKSEWCKYELDF 719
>TLR2_CRIGR (Q9R1F8) Toll-like receptor 2 precursor
Length = 784
Score = 33.5 bits (75), Expect = 0.62
Identities = 22/84 (26%), Positives = 38/84 (45%), Gaps = 5/84 (5%)
Query: 139 EINYDVFVSFRGPDTRFNFTDHLFAALQRRGINAFR---DDTKLKKGEFIAPGLFRAIEA 195
+I YD FVS+ D+ + ++L F+ G++I + +IE
Sbjct: 638 DICYDAFVSYSEQDSYW--VENLMVQQLENSEPPFKLCLHKRDFVPGKWIIDNIIDSIEK 695
Query: 196 SQLYIVVFSKNYASSTWCLRELEY 219
S + V S+N+ S WC EL++
Sbjct: 696 SHKTLFVLSENFVRSEWCKYELDF 719
>TLR2_HUMAN (O60603) Toll-like receptor 2 precursor
(Toll/interleukin 1 receptor-like protein 4)
Length = 784
Score = 32.7 bits (73), Expect = 1.1
Identities = 24/90 (26%), Positives = 36/90 (39%), Gaps = 19/90 (21%)
Query: 140 INYDVFVSFRGPDTRF----------NFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGL 189
I YD FVS+ D + NF L +R G++I +
Sbjct: 639 ICYDAFVSYSERDAYWVENLMVQELENFNPPFKLCLHKRDFIP---------GKWIIDNI 689
Query: 190 FRAIEASQLYIVVFSKNYASSTWCLRELEY 219
+IE S + V S+N+ S WC EL++
Sbjct: 690 IDSIEKSHKTVFVLSENFVKSEWCKYELDF 719
Score = 30.0 bits (66), Expect = 6.8
Identities = 15/42 (35%), Positives = 22/42 (51%)
Query: 3 DYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAY 44
D+ PG +I +IE S + V S+NF S +C EL +
Sbjct: 678 DFIPGKWIIDNIIDSIEKSHKTVFVLSENFVKSEWCKYELDF 719
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 32.3 bits (72), Expect = 1.4
Identities = 28/111 (25%), Positives = 47/111 (42%), Gaps = 17/111 (15%)
Query: 141 NYDVFVSFRGPD-TRFNFTDHLFAALQRRGINAFRDDTKLKKGEFIAPGLFRAIEASQLY 199
+YDV + + D + +F HL A+L RRGI+ + ++ A+ ++
Sbjct: 667 DYDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEVD-----------ALPKCRVL 715
Query: 200 IVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQKQSGGY 250
I+V + Y S L IL + + PIFY + P + S Y
Sbjct: 716 IIVLTSTYVPS-----NLLNILEHQHTEDRVVYPIFYRLSPYDFVCNSKNY 761
>PHS2_DICDI (P34114) Glycogen phosphorylase 2 (EC 2.4.1.1) (GP2)
Length = 992
Score = 32.0 bits (71), Expect = 1.8
Identities = 50/202 (24%), Positives = 88/202 (42%), Gaps = 31/202 (15%)
Query: 1 MDDYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILHCSVLYGKCILPIF 60
+D PGD+ R+ I++S +V + + +AY LH S L + P+F
Sbjct: 497 VDQKWPGDMSKRRALSIIDESDGKFIVMAFLAIVGAHTINGVAY-LH-SELVKHDVFPLF 554
Query: 61 YDVDPSEVRKQSGGYGESLAKLEEIAPQV----------QRWREALQLVGNISGWDLCHK 110
Y+V P++ + ++ G S + +E+ PQ+ RW L ++ DL H
Sbjct: 555 YEVWPNKFQSKTSGVTPS-SWIEQSNPQLAELITRSLNSDRWLVNLDIIK-----DLVHL 608
Query: 111 PQHAELENIIEHIN----ILGCKFSSRTKDLVEINYDVFVSFRGPDTRFN-FTDHLFAAL 165
++ + IN I K+ + D +++N DV F RF+ + L L
Sbjct: 609 ADNSSFQKEWMTINRNNKIRLAKYIEKRCD-IQVNVDVL--FDVQVKRFHEYKRQLLNVL 665
Query: 166 QRRGINAFRDDTKLKKGEFIAP 187
IN + D +K+G+ +AP
Sbjct: 666 S--VINRYLD---IKEGKKVAP 682
>NU5M_TRYBB (P04540) NADH-ubiquinone oxidoreductase chain 5 (EC
1.6.5.3)
Length = 590
Score = 31.6 bits (70), Expect = 2.3
Identities = 15/49 (30%), Positives = 25/49 (50%), Gaps = 8/49 (16%)
Query: 24 FIVVFSKNFADSCFCLVELAYIL--------HCSVLYGKCILPIFYDVD 64
+ ++F F CFCLV+ ++L +C + CIL IF+ +D
Sbjct: 427 YFLLFFLMFVFKCFCLVDCLFLLFDYECCLVYCLISLYMCILSIFFIID 475
>TOLL_DROME (P08953) Toll protein precursor
Length = 1097
Score = 30.8 bits (68), Expect = 4.0
Identities = 25/107 (23%), Positives = 45/107 (41%), Gaps = 5/107 (4%)
Query: 142 YDVFVSFRGPDTRFNFTDHLFAALQRRGINAFR---DDTKLKKGEFIAPGLFRAIEASQL 198
+D F+S+ D F D+L L+ G F+ + G I + R++ S+
Sbjct: 859 FDAFISYSHKDQSF-IEDYLVPQLEH-GPQKFQLCVHERDWLVGGHIPENIMRSVADSRR 916
Query: 199 YIVVFSKNYASSTWCLRELEYILHCSKKYGKHILPIFYDVDPSEVQK 245
I+V S+N+ S W E + G+ + + D +V+K
Sbjct: 917 TIIVLSQNFIKSEWARLEFRAAHRSALNEGRSRIIVIIYSDIGDVEK 963
>CH60_SYNP7 (P22879) 60 kDa chaperonin (Protein Cpn60) (groEL
protein)
Length = 544
Score = 30.8 bits (68), Expect = 4.0
Identities = 20/64 (31%), Positives = 31/64 (48%), Gaps = 3/64 (4%)
Query: 74 GYGESLAKLEEIAPQVQRWREALQLVGNISGWDLCHKPQHAELENIIEHINILGCKFSSR 133
G G +LA L APQ++ W A ++G + + A L+ I E+ + G S R
Sbjct: 412 GGGTTLAHL---APQLEEWATANLSGEELTGAQIVARALTARLKRIAENAGLNGAVISER 468
Query: 134 TKDL 137
K+L
Sbjct: 469 VKEL 472
>DHG_THEAC (P13203) Glucose 1-dehydrogenase (EC 1.1.1.47)
Length = 360
Score = 30.4 bits (67), Expect = 5.2
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 2/55 (3%)
Query: 104 GWDLCHKPQHAELENIIEHINILGCKFSSRTKDLVEINYDVFVSFRG-PDTRFNF 157
G+D+ +H E EN ++ ++ G KF++ KD+ E D+ V G P T F F
Sbjct: 208 GFDVTMVNRHDETENKLKIMDEFGVKFANYLKDMPE-KIDLLVDTSGDPTTTFKF 261
>LCRD_YERPE (P31487) Low calcium response locus protein D
Length = 704
Score = 30.0 bits (66), Expect = 6.8
Identities = 20/73 (27%), Positives = 38/73 (51%), Gaps = 12/73 (16%)
Query: 68 VRKQSGGYGESLAKLEEIAPQVQRWREALQLVGNISGWDLCHKPQHAELENIIEHINILG 127
+ + GGYGE + +++ I P +QR E LQ + G D+ + + LE ++E
Sbjct: 522 LEQMEGGYGELIKEVQRIVP-LQRMTEILQ---RLVGEDISIRNMRSILEAMVE------ 571
Query: 128 CKFSSRTKDLVEI 140
+ + KD+V++
Sbjct: 572 --WGQKEKDVVQL 582
>LCRD_YEREN (P21210) Low calcium response locus protein D
Length = 704
Score = 30.0 bits (66), Expect = 6.8
Identities = 20/73 (27%), Positives = 38/73 (51%), Gaps = 12/73 (16%)
Query: 68 VRKQSGGYGESLAKLEEIAPQVQRWREALQLVGNISGWDLCHKPQHAELENIIEHINILG 127
+ + GGYGE + +++ I P +QR E LQ + G D+ + + LE ++E
Sbjct: 522 LEQMEGGYGELIKEVQRIVP-LQRMTEILQ---RLVGEDISIRNMRSILEAMVE------ 571
Query: 128 CKFSSRTKDLVEI 140
+ + KD+V++
Sbjct: 572 --WGQKEKDVVQL 582
>TLR6_HUMAN (Q9Y2C9) Toll-like receptor 6 precursor
Length = 796
Score = 29.6 bits (65), Expect = 8.9
Identities = 15/45 (33%), Positives = 22/45 (48%)
Query: 3 DYHPGDLLIRKTFHAIEDSQVFIVVFSKNFADSCFCLVELAYILH 47
++ PG ++ + IE S I V S NF S +C EL + H
Sbjct: 677 NFVPGKSIVENIINCIEKSYKSIFVLSPNFVQSEWCHYELYFAHH 721
>SBCC_PSEAE (Q9HWB8) Nuclease sbcCD subunit C
Length = 1211
Score = 29.6 bits (65), Expect = 8.9
Identities = 14/47 (29%), Positives = 25/47 (52%)
Query: 79 LAKLEEIAPQVQRWREALQLVGNISGWDLCHKPQHAELENIIEHINI 125
LA+LE + +RW +L+G+ SG Q L+ +++H N+
Sbjct: 1035 LAELERARAEFRRWGRLNELIGSSSGDKFRRIAQGYNLDLLVQHSNV 1081
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.323 0.140 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,455,426
Number of Sequences: 164201
Number of extensions: 1478036
Number of successful extensions: 2914
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 2892
Number of HSP's gapped (non-prelim): 28
length of query: 287
length of database: 59,974,054
effective HSP length: 109
effective length of query: 178
effective length of database: 42,076,145
effective search space: 7489553810
effective search space used: 7489553810
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 65 (29.6 bits)
Medicago: description of AC146863.3


