

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.5 + phase: 0 /pseudo
(624 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
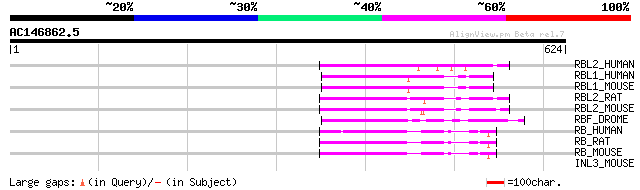
Score E
Sequences producing significant alignments: (bits) Value
RBL2_HUMAN (Q08999) Retinoblastoma-like protein 2 (130 kDa retin... 100 2e-20
RBL1_HUMAN (P28749) Retinoblastoma-like protein 1 (107 kDa retin... 98 7e-20
RBL1_MOUSE (Q64701) Retinoblastoma-like protein 1 (107 kDa retin... 97 1e-19
RBL2_RAT (O55081) Retinoblastoma-like protein 2 (130 kDa retinob... 93 2e-18
RBL2_MOUSE (Q64700) Retinoblastoma-like protein 2 (130 kDa retin... 91 7e-18
RBF_DROME (Q24472) Retinoblastoma-family protein 86 2e-16
RB_HUMAN (P06400) Retinoblastoma-associated protein (PP110) (P10... 83 2e-15
RB_RAT (P33568) Retinoblastoma-associated protein (PP105) (RB) (... 80 2e-14
RB_MOUSE (P13405) Retinoblastoma-associated protein (PP105) (RB) 80 2e-14
INL3_MOUSE (O09107) Insulin-like 3 precursor (Leydig insulin-lik... 32 6.5
>RBL2_HUMAN (Q08999) Retinoblastoma-like protein 2 (130 kDa
retinoblastoma-associated protein) (PRB2) (P130) (RBR-2)
Length = 1139
Score = 99.8 bits (247), Expect = 2e-20
Identities = 66/245 (26%), Positives = 122/245 (48%), Gaps = 34/245 (13%)
Query: 349 TGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYSLFQRILNQWTSLFFNRHIDQIIL 408
+ +S+FF K+ L AVR+ + +L +S ++R+ +++ F+ + Q L +RH+DQ+++
Sbjct: 834 SSLSLFFRKVYHLAAVRLRDLCAKLDISDELRKKIWTCFEFSIIQCPELMMDRHLDQLLM 893
Query: 409 CCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGS---------- 458
C Y +AK+++ + +F+ I+ YR QPQ + QV+RSV + RRN GS
Sbjct: 894 CAIYVMAKVTKEDKSFQNIMRCYRTQPQARSQVYRSVLIKGKRKRRNSGSSDSRSHQNSP 953
Query: 459 ----RHRTGQEHIDIISFYNEVFIP---SVKPLLVELGPGGATV----RSDQVPEANNKN 507
+ RT ++ ++ + + +P S P L + + R D + NN
Sbjct: 954 TELNKDRTSRDSSPVMRSSSTLPVPQPSSAPPTPTRLTGANSDMEEEERGDLIQFYNNIY 1013
Query: 508 DGHL--------VQNPGSPRISPFPSLPDMSPKKV--SAAHNVYVSPLRSSKMDALISHS 557
+ N +P +SP+P + SP+++ S H VY+SP K + ++S
Sbjct: 1014 IKQIKTFAMKYSQANMDAPPLSPYPFVRTGSPRRIQLSQNHPVYISP---HKNETMLSPR 1070
Query: 558 SKSYY 562
K +Y
Sbjct: 1071 EKIFY 1075
>RBL1_HUMAN (P28749) Retinoblastoma-like protein 1 (107 kDa
retinoblastoma-associated protein) (PRB1) (P107)
Length = 1068
Score = 97.8 bits (242), Expect = 7e-20
Identities = 59/220 (26%), Positives = 108/220 (48%), Gaps = 44/220 (20%)
Query: 351 ISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYSLFQRILNQWTSLFFNRHIDQIILCC 410
+++F+ K+ L +VR+ + +L +S ++R +++ F+ L L +RH+DQ++LC
Sbjct: 788 LALFYRKVYHLASVRLRDLCLKLDVSNELRRKIWTCFEFTLVHCPDLMKDRHLDQLLLCA 847
Query: 411 FYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFV----------------------- 447
FY +AK+++ TF+EI+ +YR QPQ V+RSV +
Sbjct: 848 FYIMAKVTKEERTFQEIMKSYRNQPQANSHVYRSVLLKSIPREVVAYNKNINDDFEMIDC 907
Query: 448 DWSSARRNGG-SRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNK 506
D A + S +E D+I FYN +++ VK ++ +
Sbjct: 908 DLEDATKTPDCSSGPVKEERSDLIKFYNTIYVGRVKSFALKY---------------DLA 952
Query: 507 NDGHLVQNPGSPRISPFPSLPDM--SPKKVSAAHNVYVSP 544
N H++ +P +SPFP + SP+++S H++Y+SP
Sbjct: 953 NQDHMMD---APPLSPFPHIKQQPGSPRRISQQHSIYISP 989
>RBL1_MOUSE (Q64701) Retinoblastoma-like protein 1 (107 kDa
retinoblastoma-associated protein) (PRB1) (P107)
Length = 1063
Score = 97.1 bits (240), Expect = 1e-19
Identities = 60/216 (27%), Positives = 107/216 (48%), Gaps = 40/216 (18%)
Query: 351 ISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYSLFQRILNQWTSLFFNRHIDQIILCC 410
+++F+ K+ L +VR+ + +L +S ++R +++ F+ L L +RH+DQ++LC
Sbjct: 787 LALFYRKVYHLASVRLRDLCLKLDVSNELRRKIWTCFEFTLVHCPDLMKDRHLDQLLLCA 846
Query: 411 FYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFV-------------------DWSS 451
FY +AK+++ TF+E + +YR QPQ V+RSV + D
Sbjct: 847 FYIMAKVTKEERTFQERMKSYRNQPQANSHVYRSVLLKSIPGGVVVYNGDCEMTDGDIED 906
Query: 452 ARRNGG-SRHRTGQEHIDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNKNDGH 510
A + S +E D+I FYN V++ VK ++ + N H
Sbjct: 907 ATKTPNCSSEPVKEERGDLIKFYNTVYVGRVKSFALKY---------------DLSNQDH 951
Query: 511 LVQNPGSPRISPFPSLPDM--SPKKVSAAHNVYVSP 544
++ +P +SPFP + SP+++S H++YVSP
Sbjct: 952 IMD---APPLSPFPHIKQQPGSPRRISQQHSLYVSP 984
>RBL2_RAT (O55081) Retinoblastoma-like protein 2 (130 kDa
retinoblastoma-associated protein) (PRB2) (P130) (RBR-2)
(PPAR-alpha interacting complex protein 128) (PRIC128)
Length = 1135
Score = 93.2 bits (230), Expect = 2e-18
Identities = 66/267 (24%), Positives = 123/267 (45%), Gaps = 78/267 (29%)
Query: 349 TGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYSLFQRILNQWTSLFFNRHIDQIIL 408
+ +S+FF K+ L VR+ + +L +S ++R+ +++ F+ + Q L +RH+DQ+++
Sbjct: 830 SSLSLFFRKVYYLAGVRLRDLCTKLDISDELRKKIWTCFEFSIVQCPELMMDRHLDQLLM 889
Query: 409 CCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGS-RHRTGQ--- 464
C Y +AK+++ + +F+ I+ YR QPQ + QV+RSV + RRN GS +R+ Q
Sbjct: 890 CAIYVMAKVTKEDKSFQNIMRCYRTQPQARSQVYRSVLI--KGKRRNSGSCENRSHQNSP 947
Query: 465 -----------------------------------------------EHIDIISFYNEVF 477
E D+I FYN ++
Sbjct: 948 TELNTDRASRDSSPVMRSNSTLPVPQPSSAPPTPTRLTGANSDIEEEERGDLIQFYNNIY 1007
Query: 478 IPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKV--S 535
++ ++ +AN++ D +P +SP+P + SP++V S
Sbjct: 1008 RKQIQTFAMKYS------------QANSQMD--------TPPLSPYPFVRTGSPRRVQLS 1047
Query: 536 AAHNVYVSPLRSSKMDALISHSSKSYY 562
+H +Y+SP K +A++S K +Y
Sbjct: 1048 QSHPIYISP---HKNEAMLSPREKIFY 1071
>RBL2_MOUSE (Q64700) Retinoblastoma-like protein 2 (130 kDa
retinoblastoma-associated protein) (PRB2) (P130) (RBR-2)
Length = 1135
Score = 91.3 bits (225), Expect = 7e-18
Identities = 65/267 (24%), Positives = 122/267 (45%), Gaps = 78/267 (29%)
Query: 349 TGISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYSLFQRILNQWTSLFFNRHIDQIIL 408
+ +++FF K+ L VR+ + +L +S ++R+ +++ F+ + Q T L +RH+DQ+++
Sbjct: 830 SSLALFFRKVYYLAGVRLRDLCIKLDISDELRKKIWTCFEFSIIQCTELMMDRHLDQLLM 889
Query: 409 CCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGSRHR------- 461
C Y +AK+++ + +F+ I+ YR QPQ + QV+RSV + RRN GS
Sbjct: 890 CAIYVMAKVTKEDRSFQNIMRCYRTQPQARSQVYRSVLI--KGKRRNSGSSESRSHQNSP 947
Query: 462 -------------------------------------TG-------QEHIDIISFYNEVF 477
TG +E D+I FYN ++
Sbjct: 948 TELNTDRASRDSSPVMRSNSTLPVPQPSSAPPTPTRLTGASSDVEEEERGDLIQFYNNIY 1007
Query: 478 IPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFPSLPDMSPKKV--S 535
++ ++ +AN++ D +P +SP+P + SP++V S
Sbjct: 1008 RKQIQAFAMKYS------------QANSQTD--------TPPLSPYPFVRTGSPRRVQLS 1047
Query: 536 AAHNVYVSPLRSSKMDALISHSSKSYY 562
+H +Y+SP + +A+ S K +Y
Sbjct: 1048 QSHPIYISPHNN---EAMPSPREKIFY 1071
>RBF_DROME (Q24472) Retinoblastoma-family protein
Length = 797
Score = 86.3 bits (212), Expect = 2e-16
Identities = 56/230 (24%), Positives = 110/230 (47%), Gaps = 36/230 (15%)
Query: 351 ISVFFSKIVKLGAVRISGMVERLQLSQQIRENVYSLFQRILNQWTSLFFNRHIDQIILCC 410
+ +F K+ LG +RI + L L ++ E+++ +F+ + T L +RH+DQ I+C
Sbjct: 582 LQIFLRKVYLLGWLRIQKLCSELSLCEKTPESIWHIFEHSITHETELMKDRHLDQNIMCA 641
Query: 411 FYGVAKISQL-NLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGSRHRTGQEHIDI 469
Y ++ ++ + F +I+ YR QPQ V+R VF+D +G + + DI
Sbjct: 642 IYIYIRVKRMEDPKFSDIMRAYRNQPQAVNSVYREVFID---INEDGEPKVK------DI 692
Query: 470 ISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFPSLPDM 529
I FYN ++P ++ +++ + P+ + + ++SP P
Sbjct: 693 IHFYNHTYVPLMRQFVIDY--------LNVTPDVSGR--------ASDLQLSPHPKERAA 736
Query: 530 SPKKVSAAHNVYVSPLRSSKMDALISHSSKSYYACXGESTHAYQSPXKDL 579
PKKV+ +H+++VS + +++ + S+ ++SP KDL
Sbjct: 737 QPKKVTQSHSLFVSQMSKNEIQQSPNQMVYSF----------FRSPAKDL 776
>RB_HUMAN (P06400) Retinoblastoma-associated protein (PP110)
(P105-RB) (RB)
Length = 928
Score = 82.8 bits (203), Expect = 2e-15
Identities = 57/207 (27%), Positives = 101/207 (48%), Gaps = 48/207 (23%)
Query: 349 TGISVFFSKIVKLGAVRISGMVERLQLSQ--QIRENVYSLFQRILNQWTSLFFNRHIDQI 406
T +S+F+ K+ +L +R++ + ERL LS+ ++ +++LFQ L L +RH+DQI
Sbjct: 645 TSLSLFYKKVYRLAYLRLNTLCERL-LSEHPELEHIIWTLFQHTLQNEYELMRDRHLDQI 703
Query: 407 ILCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGSRHRTGQEH 466
++C YG+ K+ ++L F+ I+ Y+ P + F+ V + +E+
Sbjct: 704 MMCSMYGICKVKNIDLKFKIIVTAYKDLPHAVQETFKRVLI--------------KEEEY 749
Query: 467 IDIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFPSL 526
II FYN VF+ +K +++ A+ R P +SP P +
Sbjct: 750 DSIIVFYNSVFMQRLKTNILQY----ASTR--------------------PPTLSPIPHI 785
Query: 527 PDMSPKKVSAA------HNVYVSPLRS 547
P SP K ++ N+Y+SPL+S
Sbjct: 786 P-RSPYKFPSSPLRIPGGNIYISPLKS 811
>RB_RAT (P33568) Retinoblastoma-associated protein (PP105) (RB)
(Fragment)
Length = 899
Score = 80.1 bits (196), Expect = 2e-14
Identities = 56/206 (27%), Positives = 97/206 (46%), Gaps = 46/206 (22%)
Query: 349 TGISVFFSKIVKLGAVRISGMVERLQLSQQIRENV-YSLFQRILNQWTSLFFNRHIDQII 407
T +S+F+ K+ +L +R++ + RL E++ ++LFQ L L +RH+DQI+
Sbjct: 616 TSLSLFYKKVYRLAYLRLNTLCARLLSDHPEPEHIIWTLFQHTLENEYELMKDRHLDQIM 675
Query: 408 LCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGSRHRTGQEHI 467
+C YG+ K+ ++L F+ I+ Y+ P + F+ V + +E
Sbjct: 676 MCSMYGICKVKNIDLKFKIIVTAYKDLPHAAQETFKRVLI--------------REEEFD 721
Query: 468 DIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFPSLP 527
II FYN VF+ +K +++ A+ R P +SP P +P
Sbjct: 722 SIIVFYNSVFMQRLKTNILQY----ASTR--------------------PPTLSPIPHIP 757
Query: 528 DMSPKKVSAA------HNVYVSPLRS 547
SP K S++ N+Y+SPL+S
Sbjct: 758 -RSPYKFSSSPLRIPGGNIYMSPLKS 782
>RB_MOUSE (P13405) Retinoblastoma-associated protein (PP105) (RB)
Length = 921
Score = 80.1 bits (196), Expect = 2e-14
Identities = 54/206 (26%), Positives = 97/206 (46%), Gaps = 46/206 (22%)
Query: 349 TGISVFFSKIVKLGAVRISGMVERLQLSQ-QIRENVYSLFQRILNQWTSLFFNRHIDQII 407
T +++F+ K+ +L +R++ + RL ++ +++LFQ L L +RH+DQI+
Sbjct: 638 TSLALFYKKVYRLAYLRLNTLCARLLSDHPELEHIIWTLFQHTLQNEYELMRDRHLDQIM 697
Query: 408 LCCFYGVAKISQLNLTFREIIYNYRKQPQCKPQVFRSVFVDWSSARRNGGSRHRTGQEHI 467
+C YG+ K+ ++L F+ I+ Y+ P + F+ V + +E
Sbjct: 698 MCSMYGICKVKNIDLKFKIIVTAYKDLPHAAQETFKRVLI--------------REEEFD 743
Query: 468 DIISFYNEVFIPSVKPLLVELGPGGATVRSDQVPEANNKNDGHLVQNPGSPRISPFPSLP 527
II FYN VF+ +K +++ A+ R P +SP P +P
Sbjct: 744 SIIVFYNSVFMQRLKTNILQY----ASTR--------------------PPTLSPIPHIP 779
Query: 528 DMSPKKVSAA------HNVYVSPLRS 547
SP K S++ N+Y+SPL+S
Sbjct: 780 -RSPYKFSSSPLRIPGGNIYISPLKS 804
>INL3_MOUSE (O09107) Insulin-like 3 precursor (Leydig insulin-like
peptide) (Ley-I-L) (Relaxin-like factor)
Length = 122
Score = 31.6 bits (70), Expect = 6.5
Identities = 22/50 (44%), Positives = 27/50 (54%), Gaps = 5/50 (10%)
Query: 483 PLLVELGPGGATVRSDQVPEANNKNDGH-----LVQNPGSPRISPFPSLP 527
PLL+ L G+ +RS Q PEA K GH LV+ G PR SP + P
Sbjct: 4 PLLLMLLALGSALRSPQPPEARAKLCGHHLVRTLVRVCGGPRWSPEATQP 53
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.342 0.148 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 61,660,444
Number of Sequences: 164201
Number of extensions: 2356009
Number of successful extensions: 7537
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 7504
Number of HSP's gapped (non-prelim): 17
length of query: 624
length of database: 59,974,054
effective HSP length: 116
effective length of query: 508
effective length of database: 40,926,738
effective search space: 20790782904
effective search space used: 20790782904
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 69 (31.2 bits)
Medicago: description of AC146862.5


