

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146862.25 + phase: 0
(206 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
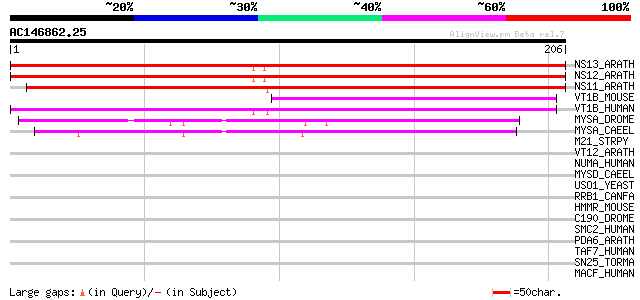
Score E
Sequences producing significant alignments: (bits) Value
NS13_ARATH (Q9LRP1) Novel plant SNARE 13 (AtNPSN13) 315 4e-86
NS12_ARATH (Q9LNH6) Novel plant SNARE 12 (AtNPSN12) 313 2e-85
NS11_ARATH (Q944A9) Novel plant SNARE 11 (AtNPSN11) 253 3e-67
VT1B_MOUSE (O88384) Vesicle transport through interaction with t... 50 3e-06
VT1B_HUMAN (Q9UEU0) Vesicle transport through interaction with t... 50 4e-06
MYSA_DROME (P05661) Myosin heavy chain, muscle 47 2e-05
MYSA_CAEEL (P12844) Myosin heavy chain A (MHC A) 45 1e-04
M21_STRPY (P50468) M protein, serotype 2.1 precursor 42 0.001
VT12_ARATH (Q9SEL5) Vesicle transport v-SNARE 12 (AtVTI12) (Vesi... 40 0.003
NUMA_HUMAN (Q14980) Nuclear mitotic apparatus protein 1 (NuMA pr... 40 0.003
MYSD_CAEEL (P02567) Myosin heavy chain D (MHC D) 40 0.005
USO1_YEAST (P25386) Intracellular protein transport protein USO1 39 0.006
RRB1_CANFA (Q28298) Ribosome-binding protein 1 (180 kDa ribosome... 39 0.006
HMMR_MOUSE (Q00547) Hyaluronan mediated motility receptor (Intra... 39 0.008
C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 1... 39 0.008
SMC2_HUMAN (O95347) Structural maintenance of chromosome 2-like ... 39 0.011
PDA6_ARATH (O22263) Probable protein disulfide-isomerase A6 prec... 39 0.011
TAF7_HUMAN (Q15545) Transcription initiation factor TFIID subuni... 38 0.019
SN25_TORMA (P36976) Synaptosomal-associated protein 25 (SNAP-25) 38 0.019
MACF_HUMAN (Q9UPN3) Microtubule-actin crosslinking factor 1, iso... 38 0.019
>NS13_ARATH (Q9LRP1) Novel plant SNARE 13 (AtNPSN13)
Length = 269
Score = 315 bits (808), Expect = 4e-86
Identities = 172/237 (72%), Positives = 188/237 (78%), Gaps = 31/237 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MASNL M+PQLEQIHGEIRD FRALANGFQ+LDKIKDS RQS QLEELT KMR+CKRL+K
Sbjct: 1 MASNLPMSPQLEQIHGEIRDHFRALANGFQRLDKIKDSTRQSKQLEELTDKMRECKRLVK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDRE+KDE A N EVNKQLNDEKQSM E NS
Sbjct: 61 EFDRELKDEEARNSPEVNKQLNDEKQSMIKELNSYVALRKTYMSTLGNKKVELFDMGAGV 120
Query: 94 ----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQ 149
TAE NVQ+AS MSNQELV+AGMK MDETDQAIERSKQVV QT+EVGTQTA+ LKGQ
Sbjct: 121 SGEPTAEENVQVASSMSNQELVDAGMKRMDETDQAIERSKQVVEQTLEVGTQTAANLKGQ 180
Query: 150 TEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
T+QMGR+VN LD+IQFSIKKASQLVKEIGRQVATDKCIM FLFLIVCGV+AII+VK+
Sbjct: 181 TDQMGRVVNHLDTIQFSIKKASQLVKEIGRQVATDKCIMGFLFLIVCGVVAIIIVKI 237
>NS12_ARATH (Q9LNH6) Novel plant SNARE 12 (AtNPSN12)
Length = 265
Score = 313 bits (801), Expect = 2e-85
Identities = 170/237 (71%), Positives = 187/237 (78%), Gaps = 31/237 (13%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS L M+P LEQIHGEIRD FRALANGFQ+LDKIKDS+RQS QLEEL KMRDCKRL+K
Sbjct: 1 MASELPMSPHLEQIHGEIRDHFRALANGFQRLDKIKDSSRQSKQLEELAEKMRDCKRLVK 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT-EKNS-------------------------- 93
+FDRE+KD A N +VNKQLNDEKQSM E NS
Sbjct: 61 EFDRELKDGEARNSPQVNKQLNDEKQSMIKELNSYVALRKTYLNTLGNKKVELFDTGAGV 120
Query: 94 ----TAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQ 149
TAE NVQ+AS MSNQELV+AGMK MDETDQAIERSKQVVHQT+EVGTQTAS LKGQ
Sbjct: 121 SGEPTAEENVQMASTMSNQELVDAGMKRMDETDQAIERSKQVVHQTLEVGTQTASNLKGQ 180
Query: 150 TEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
T+QMGR+VN+LD+IQFS+KKASQLVKEIGRQVATDKCIM FLFLIVCGVIAII+VK+
Sbjct: 181 TDQMGRVVNDLDTIQFSLKKASQLVKEIGRQVATDKCIMAFLFLIVCGVIAIIIVKI 237
>NS11_ARATH (Q944A9) Novel plant SNARE 11 (AtNPSN11)
Length = 265
Score = 253 bits (645), Expect = 3e-67
Identities = 136/229 (59%), Positives = 164/229 (71%), Gaps = 29/229 (12%)
Query: 7 MTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREI 66
++ +L +I G+I DIFRAL+NGFQKL+KIKD+NRQS QLEELT KMRDCK LIKDFDREI
Sbjct: 7 VSEELAEIEGQINDIFRALSNGFQKLEKIKDANRQSRQLEELTDKMRDCKSLIKDFDREI 66
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKNST-----------------------------AEG 97
K +GN N+ LND +QSM ++ ++ E
Sbjct: 67 KSLESGNDASTNRMLNDRRQSMVKELNSYVALKKKYSSNLASNNKRVDLFDGPGEEHMEE 126
Query: 98 NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIV 157
NV LAS MSNQEL++ G MD+TDQAIER K++V +TI VGT T++ LK QTEQM R+V
Sbjct: 127 NVLLASNMSNQELMDKGNSMMDDTDQAIERGKKIVQETINVGTDTSAALKPQTEQMSRVV 186
Query: 158 NELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIVVKV 206
NELDSI FS+KKAS+LVKEIGRQVATDKCIM FLFLIV GVIAII+VK+
Sbjct: 187 NELDSIHFSLKKASKLVKEIGRQVATDKCIMAFLFLIVIGVIAIIIVKI 235
>VT1B_MOUSE (O88384) Vesicle transport through interaction with
t-SNAREs homolog 1B (Vesicle transport v-SNARE protein
Vti1-like 1) (Vti1-rp1)
Length = 232
Score = 50.4 bits (119), Expect = 3e-06
Identities = 25/106 (23%), Positives = 59/106 (55%)
Query: 98 NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIV 157
N L S + L+ G ++++ Q+IERS ++ +T ++GT+ L Q +Q+ R
Sbjct: 119 NEHLNRLQSQRALLLQGTESLNRATQSIERSHRIATETDQIGTEIIEELGEQRDQLERTK 178
Query: 158 NELDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIV 203
+ L + ++ K+ ++++ + R+V T+K ++ + L+ ++ +V
Sbjct: 179 SRLVNTNENLSKSRKILRSMSRKVITNKLLLSVIILLELAILVGLV 224
>VT1B_HUMAN (Q9UEU0) Vesicle transport through interaction with
t-SNAREs homolog 1B (Vesicle transport v-SNARE protein
Vti1-like 1) (Vti1-rp1)
Length = 232
Score = 50.1 bits (118), Expect = 4e-06
Identities = 37/224 (16%), Positives = 98/224 (43%), Gaps = 21/224 (9%)
Query: 1 MASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIK 60
MAS+ + E++H R + L ++L + + + + K ++ +
Sbjct: 1 MASSAASSEHFEKLHEIFRGLHENLQGVPERLLGTAGTEEKKKLIRDFDEKQQEANETLA 60
Query: 61 DFDREIKDEGAGNPEEVNKQLNDEKQSMT----EKNST-----------------AEGNV 99
+ + E++ + +L + ++ + E ST A N
Sbjct: 61 EMEEELRYAPLSFRNPMMSKLRNYRKDLAKLHREVRSTPLTATPGGRGDMKYGIYAVENE 120
Query: 100 QLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNE 159
+ S + ++ G ++++ Q+IERS ++ +T ++G++ L Q +Q+ R +
Sbjct: 121 HMNRLQSQRAMLLQGTESLNRATQSIERSHRIATETDQIGSEIIEELGEQRDQLERTKSR 180
Query: 160 LDSIQFSIKKASQLVKEIGRQVATDKCIMLFLFLIVCGVIAIIV 203
L + ++ K+ ++++ + R+V T+K ++ + L+ ++ +V
Sbjct: 181 LVNTSENLSKSRKILRSMSRKVTTNKLLLSIIILLELAILGGLV 224
>MYSA_DROME (P05661) Myosin heavy chain, muscle
Length = 1962
Score = 47.4 bits (111), Expect = 2e-05
Identities = 46/201 (22%), Positives = 89/201 (43%), Gaps = 18/201 (8%)
Query: 4 NLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRL----- 58
N K+T Q + ++RDI L +++ +++ Q E++G +D + L
Sbjct: 904 NAKLTAQKNDLENQLRDIQERLTQEEDARNQLFQQKKKADQ--EISGLKKDIEDLELNVQ 961
Query: 59 -------IKDFD-REIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQ-E 109
KD R + DE A E +NK LN EK+ E N +Q A + N
Sbjct: 962 KAEQDKATKDHQIRNLNDEIAHQDELINK-LNKEKKMQGETNQKTGEELQAAEDKINHLN 1020
Query: 110 LVNAGMK-TMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIK 168
V A ++ T+DE + ++ER K+V + + LK E + + ++ +I+
Sbjct: 1021 KVKAKLEQTLDELEDSLEREKKVRGDVEKSKRKVEGDLKLTQEAVADLERNKKELEQTIQ 1080
Query: 169 KASQLVKEIGRQVATDKCIML 189
+ + + I ++ ++ ++L
Sbjct: 1081 RKDKELSSITAKLEDEQVVVL 1101
>MYSA_CAEEL (P12844) Myosin heavy chain A (MHC A)
Length = 1969
Score = 45.1 bits (105), Expect = 1e-04
Identities = 41/185 (22%), Positives = 85/185 (45%), Gaps = 7/185 (3%)
Query: 10 QLEQIHGEIRDIFRA---LANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFD-RE 65
+L I G++ D+ LA +K D+ ++ Q EL+ + + ++ +D + R
Sbjct: 931 KLSDITGQLEDMQERNEDLARQKKKTDQELSDTKKHVQDLELSLRKAEQEKQSRDHNIRS 990
Query: 66 IKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSN--QELVNAGMKTMDETDQ 123
++DE A E V K LN EK+ E N ++Q + N +++ N + MDE ++
Sbjct: 991 LQDEMANQDEAVAK-LNKEKKHQEESNRKLNEDLQSEEDKVNHLEKIRNKLEQQMDELEE 1049
Query: 124 AIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVAT 183
I+R K+ + + LK E + I + ++ ++K+ + + ++A
Sbjct: 1050 NIDREKRSRGDIEKAKRKVEGDLKVAQENIDEITKQKHDVETTLKRKEEDLHHTNAKLAE 1109
Query: 184 DKCIM 188
+ I+
Sbjct: 1110 NNSII 1114
>M21_STRPY (P50468) M protein, serotype 2.1 precursor
Length = 407
Score = 41.6 bits (96), Expect = 0.001
Identities = 38/180 (21%), Positives = 81/180 (44%), Gaps = 15/180 (8%)
Query: 13 QIHGEIRDIFRALANGFQKLDKIKDSNRQ-----STQLEELTGKMRDCKR-LIKDFDREI 66
++H +I+++ A F+KLDK+++ +++ E+L K D +R ++ D+E
Sbjct: 59 ELHDKIKNLEEEKAELFEKLDKVEEEHKKVEEEHKKDHEKLEKKSEDVERHYLRQLDQEY 118
Query: 67 KD--EGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQA 124
K+ E N EE+ +Q E + ++ + QL E E ++ E +A
Sbjct: 119 KEQQERQKNLEELERQSQREVEKRYQEQ--LQKQQQLEKEKQISEASRKSLRRDLEASRA 176
Query: 125 IERSKQVVHQTI----EVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQ 180
++ + HQ + ++ + +L+ E +L++ +K+ Q + E RQ
Sbjct: 177 AKKDLEAEHQKLKEEKQISEASRKSLRRDLEASRAAKKDLEAEHQKLKEEKQ-ISEASRQ 235
>VT12_ARATH (Q9SEL5) Vesicle transport v-SNARE 12 (AtVTI12) (Vesicle
transport v-SNARE protein VTI1b) (Vesicle soluble NSF
attachment protein receptor VTI1b) (AtVTI1b)
Length = 223
Score = 40.4 bits (93), Expect = 0.003
Identities = 42/199 (21%), Positives = 85/199 (42%), Gaps = 30/199 (15%)
Query: 25 LANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDE 84
L+NG +K KI E++ + + LI+ D E + +L +
Sbjct: 29 LSNGEEKKGKIA---------EQIKSGIDEADVLIRKMDLEARSLQPSAKAVCLSKLREY 79
Query: 85 KQSMTE-KNSTAEGNVQLASEMSNQELVNAGM------------------KTMDETDQAI 125
K + + K + A S +EL+ +GM + +D++ I
Sbjct: 80 KSDLNQLKKEFKRVSSADAKPSSREELMESGMADLHAVSADQRGRLAMSVERLDQSSDRI 139
Query: 126 ERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGRQVATDK 185
S++++ +T EVG L Q + + N+L + +I K+ +++ + R++ +K
Sbjct: 140 RESRRLMLETEEVGISIVQDLSQQRQTLLHAHNKLHGVDDAIDKSKKVLTAMSRRMTRNK 199
Query: 186 CIMLFLFLIVCGVIAIIVV 204
I+ +IV V+AII++
Sbjct: 200 WII--TSVIVALVLAIILI 216
>NUMA_HUMAN (Q14980) Nuclear mitotic apparatus protein 1 (NuMA
protein) (SP-H antigen)
Length = 2115
Score = 40.4 bits (93), Expect = 0.003
Identities = 45/178 (25%), Positives = 78/178 (43%), Gaps = 16/178 (8%)
Query: 10 QLEQIHGEIRDIFRALANGFQKLDKIKDSNRQS-TQLE----ELTGKMRDCKRLIKDFDR 64
QLE + E L Q ++ +DS + S TQ + EL+ K+ + + ++ +
Sbjct: 605 QLEALEKEKAAKLEILQQQLQVANEARDSAQTSVTQAQREKAELSRKVEELQACVETARQ 664
Query: 65 EIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQA 124
E + E E+ QL E+Q TEK A+ QL QE + A +++ T +
Sbjct: 665 E-QHEAQAQVAELELQLRSEQQKATEKERVAQEKDQL------QEQLQALKESLKVTKGS 717
Query: 125 IERSKQVVHQTIEVGTQTASTLKGQT----EQMGRIVNELDSIQFSIKKASQLVKEIG 178
+E K+ +E + S LK +T EQ R EL+ + K ++++G
Sbjct: 718 LEEEKRRAADALEEQQRCISELKAETRSLVEQHKRERKELEEERAGRKGLEARLQQLG 775
Score = 33.5 bits (75), Expect = 0.36
Identities = 32/107 (29%), Positives = 54/107 (49%), Gaps = 11/107 (10%)
Query: 81 LNDEKQSMTEKNSTAEG---NVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIE 137
L+ K+ + E+ S A G Q+AS S +NA T+ + DQ + KQ Q E
Sbjct: 478 LSQAKEEL-EQASQAHGARLTAQVASLTSELTTLNA---TIQQQDQELAGLKQ---QAKE 530
Query: 138 VGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIG-RQVAT 183
Q A TL+ Q + + ++++ + S+K+ Q +KE+ +Q AT
Sbjct: 531 KQAQLAQTLQQQEQASQGLRHQVEQLSSSLKQKEQQLKEVAEKQEAT 577
>MYSD_CAEEL (P02567) Myosin heavy chain D (MHC D)
Length = 1938
Score = 39.7 bits (91), Expect = 0.005
Identities = 31/128 (24%), Positives = 62/128 (48%), Gaps = 11/128 (8%)
Query: 66 IKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQL--ASEMSNQELVNAGMKTMDETDQ 123
++DE E + K +N EK+ + E N ++Q A + L +T+DE ++
Sbjct: 985 LQDEMNSQDETIGK-INKEKKLLEENNRQLVDDLQAEEAKQAQANRLRGKLEQTLDEMEE 1043
Query: 124 AIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQF----SIKKASQLVKEIGR 179
A+ER K++ +T + + LKG E ++EL +I+ S+KK + +G
Sbjct: 1044 AVEREKRIRAETEKSKRKVEGELKGAQE----TIDELSAIKLETDASLKKKEADIHALGV 1099
Query: 180 QVATDKCI 187
++ ++ +
Sbjct: 1100 RIEDEQAL 1107
>USO1_YEAST (P25386) Intracellular protein transport protein USO1
Length = 1790
Score = 39.3 bits (90), Expect = 0.006
Identities = 35/165 (21%), Positives = 77/165 (46%), Gaps = 15/165 (9%)
Query: 11 LEQIHGEIRDIFRALANGFQKLDKI-KDSNRQSTQLEELTGKMRDCKRLIKDFDREIKDE 69
++ + EI + +KL I +D+ R L+E ++ K +++ +++++E
Sbjct: 1433 IKSLQDEILSYKDKITRNDEKLLSIERDNKRDLESLKEQLRAAQESKAKVEEGLKKLEEE 1492
Query: 70 GAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSK 129
+ E+ K K+ M + ST E SN+ + + M+T+ ++D+ +E+SK
Sbjct: 1493 SSKEKAELEKS----KEMMKKLESTIE---------SNETELKSSMETIRKSDEKLEQSK 1539
Query: 130 QVVHQTIEVGTQTASTLKGQTEQMGRIVNELDS-IQFSIKKASQL 173
+ + I+ S L + + + + EL S ++ K S+L
Sbjct: 1540 KSAEEDIKNLQHEKSDLISRINESEKDIEELKSKLRIEAKSGSEL 1584
Score = 35.0 bits (79), Expect = 0.12
Identities = 35/142 (24%), Positives = 64/142 (44%), Gaps = 12/142 (8%)
Query: 39 NRQSTQLEELTGKMRDCKRLIKDFDREIKDEGAGNPEEVNKQLNDE-------KQSMTEK 91
N LE L + D +K ++ +I ++ EE++ QLNDE +S+ +K
Sbjct: 1141 NSLRANLESLEKEHEDLAAQLKKYEEQIANKERQYNEEIS-QLNDEITSTQQENESIKKK 1199
Query: 92 NSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTE 151
N EG V+ A + +++E N +D + I+ K+ ++T E ++K
Sbjct: 1200 NDELEGEVK-AMKSTSEEQSNLKKSEIDALNLQIKELKK-KNETNE--ASLLESIKSVES 1255
Query: 152 QMGRIVNELDSIQFSIKKASQL 173
+ +I D F K+ S+L
Sbjct: 1256 ETVKIKELQDECNFKEKEVSEL 1277
>RRB1_CANFA (Q28298) Ribosome-binding protein 1 (180 kDa ribosome
receptor) (RRp)
Length = 1534
Score = 39.3 bits (90), Expect = 0.006
Identities = 40/179 (22%), Positives = 84/179 (46%), Gaps = 16/179 (8%)
Query: 2 ASNLKMTPQLEQIHGEIRDIFRALANG--------FQKLDKIKDSNRQSTQLEELTGKMR 53
AS + +++Q+ G+IR + L NG Q+ ++D+ Q+T E + +
Sbjct: 896 ASYREHVKEVQQLQGKIRTLQEQLENGPNTQLARLQQENSILRDALNQATSQVE-SKQNT 954
Query: 54 DCKRLIKDFDREIKDEGAGNPEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNA 113
+ +L ++ + K+ E +Q +++++ K + E V L + S++E A
Sbjct: 955 ELAKLRQELSKVSKE--LVEKSEAARQEEQQRKALETKTAALEKQV-LQLQASHKESEEA 1011
Query: 114 GMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQ 172
K +DE + + RS Q H ++ + A + Q +QM + ++L S + +K S+
Sbjct: 1012 LQKRLDEVSRELCRS-QTSHASLRADAEKA---QEQQQQMAELHSKLQSSEAEVKSKSE 1066
>HMMR_MOUSE (Q00547) Hyaluronan mediated motility receptor
(Intracellular hyaluronic acid binding protein)
(Receptor for hyaluronan-mediated motility)
Length = 794
Score = 38.9 bits (89), Expect = 0.008
Identities = 34/149 (22%), Positives = 66/149 (43%), Gaps = 8/149 (5%)
Query: 18 IRDIFRALANGFQKLDKIKDSNRQST-QLEELTGKMRDCKRLIKDFDREIKDEGAGNPEE 76
+RD+ L + +K + S R T QLE + K D + ++D +++ + +E
Sbjct: 494 LRDVTAQLESEQEKYNDTAQSLRDVTAQLESVQEKYNDTAQSLRDVSAQLESYKSSTLKE 553
Query: 77 VNKQLNDEKQSMTEKNSTAEGNVQ------LASEMSNQELVNAGMKTMDETDQAIERSKQ 130
+ + L E ++ EK + AE +V+ L +E +NQE + + E K+
Sbjct: 554 I-EDLKLENLTLQEKVAMAEKSVEDVQQQILTAESTNQEYARMVQDLQNRSTLKEEEIKE 612
Query: 131 VVHQTIEVGTQTASTLKGQTEQMGRIVNE 159
+ +E T + L+ Q E + + E
Sbjct: 613 ITSSFLEKITDLKNQLRQQDEDFRKQLEE 641
>C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 190)
(Microtubule binding protein 190) (d-CLIP-190)
Length = 1690
Score = 38.9 bits (89), Expect = 0.008
Identities = 42/193 (21%), Positives = 83/193 (42%), Gaps = 42/193 (21%)
Query: 33 DKIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKD----------------EGAGNPEE 76
D++ +S ++ QL+E K+ + +++ + +IKD + A + +
Sbjct: 1280 DQLLESQKKEKQLQEEAAKLSGELQQVQEANGDIKDSLVKVEELVKVLEEKLQAATSQLD 1339
Query: 77 VNKQLNDEKQSMTEKNSTAEGNVQLAS----------EMSNQELVNA------GMK---- 116
+ N E Q + K+ EGN+Q S E +N EL A G+K
Sbjct: 1340 AQQATNKELQELLVKSQENEGNLQGESLAVTEKLQQLEQANGELKEALCQKENGLKELQG 1399
Query: 117 TMDETDQAIE---RSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKA--- 170
+DE++ +E +S + +E Q TL+ +T ++ +++L ++K+
Sbjct: 1400 KLDESNTVLESQKKSHNEIQDKLEQAQQKERTLQEETSKLAEQLSQLKQANEELQKSLQQ 1459
Query: 171 SQLVKEIGRQVAT 183
QL+ E G + T
Sbjct: 1460 KQLLLEKGNEFDT 1472
Score = 37.4 bits (85), Expect = 0.025
Identities = 30/129 (23%), Positives = 64/129 (49%), Gaps = 10/129 (7%)
Query: 10 QLEQIHGEIRDIFRALANGFQKLD-KIKDSNRQSTQLEELTGKMRDCKRLIKDFDREIKD 68
QLEQ +GE+++ NG ++L K+ +SN ++ +++D + +R +++
Sbjct: 1375 QLEQANGELKEALCQKENGLKELQGKLDESNTVLESQKKSHNEIQDKLEQAQQKERTLQE 1434
Query: 69 EGAGNPEEVN--KQLNDEKQ-SMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAI 125
E + E+++ KQ N+E Q S+ +K E + ++++ + K +DE D A
Sbjct: 1435 ETSKLAEQLSQLKQANEELQKSLQQKQLLLEKGNEFDTQLAEYQ------KVIDEMDDAA 1488
Query: 126 ERSKQVVHQ 134
++ Q
Sbjct: 1489 SVKSALLEQ 1497
Score = 32.0 bits (71), Expect = 1.0
Identities = 35/186 (18%), Positives = 84/186 (44%), Gaps = 13/186 (6%)
Query: 6 KMTPQLEQI---HGEIRDIFRALANGFQKLDKIKDSNRQSTQLEELTGKMRDCKRLIKDF 62
K+ QLEQ H +++ L +K IK+ ++ QL+ + + ++++
Sbjct: 780 KLQQQLEQKTLGHEKLQAALEELKK--EKETIIKEKEQELQQLQSKSAESESALKVVQVQ 837
Query: 63 DREIKDEGAGNPEEVNK---QLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMD 119
+++ + A + EE +K +L+DE + K+ E +L S SN E A K ++
Sbjct: 838 LEQLQQQAAASGEEGSKTVAKLHDEISQL--KSQAEETQSELKSTQSNLE---AKSKQLE 892
Query: 120 ETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKEIGR 179
+ ++E + +E T+ S + + +++S ++ A+ ++++ +
Sbjct: 893 AANGSLEEEAKKSGHLLEQITKLKSEVGETQAALSSCHTDVESKTKQLEAANAALEKVNK 952
Query: 180 QVATDK 185
+ A +
Sbjct: 953 EYAESR 958
Score = 28.9 bits (63), Expect = 8.8
Identities = 44/204 (21%), Positives = 86/204 (41%), Gaps = 25/204 (12%)
Query: 2 ASNLKMTPQLEQIHGEIRDIFRALANGFQKLDKIKDS-NRQSTQLEELTGKMRDCKR--- 57
+ + T + E + E+R + Q+LD++ N Q L +R K
Sbjct: 608 SERVNKTDECEILQTEVRMRDEQIRELNQQLDEVTTQLNVQKADSSALDDMLRLQKEGTE 667
Query: 58 ----LIKDFDREI---KDEGA---GNPEEVNKQLNDEKQS------MTEKNSTAEGNVQL 101
L++ ++E+ K++ A + E++ KQ++D KQ + E A +QL
Sbjct: 668 EKSTLLEKTEKELVQSKEQAAKTLNDKEQLEKQISDLKQLAEQEKLVREMTENAINQIQL 727
Query: 102 ASEMSNQELVNAGMKTMDETDQAIERSKQVVH-QTIEV-GTQTASTLKGQTEQMGRIVNE 159
E Q+L +K + D ++S+ VH Q I+ TQ L E + ++ +
Sbjct: 728 EKESIEQQL---ALKQNELEDFQKKQSESEVHLQEIKAQNTQKDFELVESGESLKKLQQQ 784
Query: 160 LDSIQFSIKKASQLVKEIGRQVAT 183
L+ +K ++E+ ++ T
Sbjct: 785 LEQKTLGHEKLQAALEELKKEKET 808
>SMC2_HUMAN (O95347) Structural maintenance of chromosome 2-like 1
protein (Chromosome-associated protein E) (hCAP-E)
(XCAP-E homolog) (PRO0324)
Length = 1197
Score = 38.5 bits (88), Expect = 0.011
Identities = 33/145 (22%), Positives = 74/145 (50%), Gaps = 7/145 (4%)
Query: 43 TQLEELTGKMRDCKRLIKDFDREIKDEGAG--NPEEVNKQLNDEKQSMTEKNSTAEGNVQ 100
T+ +EL ++D R+ ++ R +++E AG N E +QL + + TE+ + +Q
Sbjct: 676 TKFQELKD-VQDELRIKENELRALEEELAGLKNTAEKYRQLKQQWEMKTEEADLLQTKLQ 734
Query: 101 LASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNE- 159
+S QE ++A KT++E+++ ++ +K++ + E + +K + R + +
Sbjct: 735 QSSYHKQQEELDALKKTIEESEETLKNTKEIQRKAEEKYEVLENKMKNAEAERERELKDA 794
Query: 160 ---LDSIQFSIKKASQLVKEIGRQV 181
LD + +S+ +KE ++V
Sbjct: 795 QKKLDCAKTKADASSKKMKEKQQEV 819
>PDA6_ARATH (O22263) Probable protein disulfide-isomerase A6
precursor (EC 5.3.4.1) (P5)
Length = 361
Score = 38.5 bits (88), Expect = 0.011
Identities = 37/132 (28%), Positives = 64/132 (48%), Gaps = 13/132 (9%)
Query: 67 KDEGAGNPEEVNKQLNDEKQSMTEKNSTA-EGNVQLASEMSNQELVNAGMKTMDETDQAI 125
KD AG+ + + L+D + EK+ T+ + QL S+ E ++A +K E A
Sbjct: 224 KDNKAGHDYDGGRDLDDFVSFINEKSGTSRDSKGQLTSKAGIVESLDALVK---ELVAAS 280
Query: 126 ERSKQVVHQTIEVGTQTASTLKGQTEQMGRIVNEL------DSIQFSIKKASQLVKEIGR 179
E K+ V IE + ASTLKG T + G++ +L ++ K+ +L + +G+
Sbjct: 281 EDEKKAVLSRIE---EEASTLKGSTTRYGKLYLKLAKSYIEKGSDYASKETERLGRVLGK 337
Query: 180 QVATDKCIMLFL 191
++ K L L
Sbjct: 338 SISPVKADELTL 349
>TAF7_HUMAN (Q15545) Transcription initiation factor TFIID subunit 7
(Transcription initiation factor TFIID 55 kDa subunit)
(TAF(II)55) (TAFII-55) (TAFII55)
Length = 349
Score = 37.7 bits (86), Expect = 0.019
Identities = 27/121 (22%), Positives = 58/121 (47%), Gaps = 7/121 (5%)
Query: 15 HGEIRDIFRALANGFQKLDKIKDSNRQSTQL----EELTGKMRDCKRLIKDFDREIKDEG 70
H E+R+IF L++ + D+ + + + + E+L +++D +L + ++ ++EG
Sbjct: 216 HDELREIFNDLSSSSEDEDETQHQDEEDINIIDTEEDLERQLQD--KLNESDEQHQENEG 273
Query: 71 AGN-PEEVNKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSK 129
+ KQ+++ K + E A+ L ++ N L N +DE Q +R K
Sbjct: 274 TNQLVMGIQKQIDNMKGKLQETQDRAKRQEDLIMKVENLALKNRFQAVLDELKQKEDREK 333
Query: 130 Q 130
+
Sbjct: 334 E 334
>SN25_TORMA (P36976) Synaptosomal-associated protein 25 (SNAP-25)
Length = 210
Score = 37.7 bits (86), Expect = 0.019
Identities = 23/94 (24%), Positives = 47/94 (49%), Gaps = 2/94 (2%)
Query: 93 STAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIEVGTQTASTLKGQTEQ 152
++ E ++ SE +E+ + DE+ ++ R Q+V ++ + G +T L Q EQ
Sbjct: 3 NSVENSMDPRSEQ--EEMQRCADQITDESLESTRRMLQLVEESKDAGIRTLVMLDEQGEQ 60
Query: 153 MGRIVNELDSIQFSIKKASQLVKEIGRQVATDKC 186
+ RI +D I +K+A + + ++G+ C
Sbjct: 61 LERIEEGMDQINKDMKEAEKNLSDLGKCCGLCSC 94
>MACF_HUMAN (Q9UPN3) Microtubule-actin crosslinking factor 1, isoforms
1/2/3 (Actin cross-linking family protein 7) (Macrophin
1) (Trabeculin-alpha) (620 kDa actin-binding protein)
(ABP620)
Length = 5430
Score = 37.7 bits (86), Expect = 0.019
Identities = 29/99 (29%), Positives = 51/99 (51%), Gaps = 6/99 (6%)
Query: 78 NKQLNDEKQSMTEKNSTAEGNVQLASEMSNQELVNAGMKTMDETDQAIERSKQVVHQTIE 137
++QLN+ Q + T G+V L++ +EL + K ++ TD+ RS Q + Q I
Sbjct: 2567 HEQLNEAAQGIL----TGPGDVSLSTSQVQKELQSINQKWVELTDKLNSRSSQ-IDQAIV 2621
Query: 138 VGTQTASTLKGQTEQMGRIVNELDSIQFSIKKASQLVKE 176
TQ L+ +E++ R V + S+Q +I + VK+
Sbjct: 2622 KSTQYQELLQDLSEKV-RAVGQRLSVQSAISTQPEAVKQ 2659
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.130 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,168,759
Number of Sequences: 164201
Number of extensions: 760401
Number of successful extensions: 3701
Number of sequences better than 10.0: 304
Number of HSP's better than 10.0 without gapping: 54
Number of HSP's successfully gapped in prelim test: 251
Number of HSP's that attempted gapping in prelim test: 3484
Number of HSP's gapped (non-prelim): 439
length of query: 206
length of database: 59,974,054
effective HSP length: 105
effective length of query: 101
effective length of database: 42,732,949
effective search space: 4316027849
effective search space used: 4316027849
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC146862.25


