

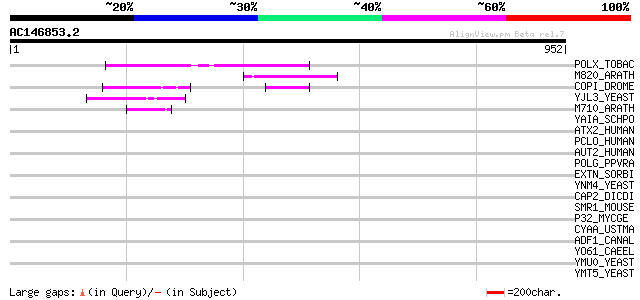
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146853.2 - phase: 0 /pseudo
(952 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 143 2e-33
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 112 5e-24
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 91 1e-17
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 52 8e-06
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 50 4e-05
YAIA_SCHPO (Q09897) Hypothetical protein C24B11.10c in chromosome I 42 0.006
ATX2_HUMAN (Q99700) Ataxin-2 (Spinocerebellar ataxia type 2 prot... 39 0.050
PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin) 37 0.19
AUT2_HUMAN (Q8WXX7) Autism susceptibility gene 2 protein 37 0.19
POLG_PPVRA (P17767) Genome polyprotein [Contains: P1 proteinase ... 37 0.25
EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein) 37 0.25
YNM4_YEAST (P53919) Hypothetical 54.9 kDa protein in SPC98-TOM70... 36 0.43
CAP2_DICDI (P34122) CABP1-related protein p31/p34 36 0.43
SMR1_MOUSE (Q61900) Submaxillary gland androgen regulated protei... 36 0.56
P32_MYCGE (Q49417) P32 adhesin (Cytadhesin P32) 36 0.56
CYAA_USTMA (P49606) Adenylate cyclase (EC 4.6.1.1) (ATP pyrophos... 36 0.56
ADF1_CANAL (P46589) Adherence factor (Adhesion and aggregation m... 36 0.56
YO61_CAEEL (P34600) Hypothetical protein ZK1098.1 in chromosome III 35 0.73
YMU0_YEAST (Q04670) Transposon Ty1 protein B 35 0.73
YMT5_YEAST (Q04214) Transposon Ty1 protein B 35 0.73
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 143 bits (361), Expect = 2e-33
Identities = 101/356 (28%), Positives = 161/356 (44%), Gaps = 23/356 (6%)
Query: 164 FQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEMATY 223
F++ C ++GI + P T NG +ER R+ +R++L A LP SFW A++ A Y
Sbjct: 560 FEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACY 619
Query: 224 LMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTKCVFLGY 283
L+N P + FE P +V +K+ SY+HL+VFGC F P + KL +S C+F+GY
Sbjct: 620 LINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGY 679
Query: 284 PSNHRGYKCLGLLSNKIIICRHVLFNENIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMD 343
GY+ + K+I R V+F E+ + T + ++ II + +
Sbjct: 680 GDEEFGYRLWDPVKKKVIRSRDVVFRES----------EVRTAADMSEKVKNGIIPNFV- 728
Query: 344 QTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQS--NPTSPIQQNLPPIFQPTL-QANTK 400
IP ++ P S P + + + + PT + +
Sbjct: 729 ------TIPSTSNNPTSAESTTDEVSEQGEQPGEVIEQGEQLDEGVEEVEHPTQGEEQHQ 782
Query: 401 PITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKM---AMNDEYNSLIKNKT 457
P+ RS+ + + + ++ P + L P AM +E SL KN T
Sbjct: 783 PLRRSERPRVESRRYPSTEYVLISDDREPESLKEVLSHPEKNQLMKAMQEEMESLQKNGT 842
Query: 458 WDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM 513
+ LV P ++ W+F+ K+ D R+KARLV G Q+ GID E FSP+
Sbjct: 843 YKLVELPKGKRPLKCKWVFKLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPV 898
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 112 bits (280), Expect = 5e-24
Identities = 67/162 (41%), Positives = 93/162 (57%), Gaps = 2/162 (1%)
Query: 402 ITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLV 461
+TRS+ GI K N KY L T P++ + ALKDP W AM +E ++L +NKTW LV
Sbjct: 2 LTRSKAGINKLNPKY-SLTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTWILV 60
Query: 462 PRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAGQQVGIDCGETFSPM*NQLLFAQ 521
P P + N++ W+F+ K SDGT +R KARLV G Q+ GI ET+SP+
Sbjct: 61 PPPVNQNILGCKWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRT 120
Query: 522 FSELLHP-DLGKFISWMSRMLFFMVN*KKQCTCTNLWDLKIL 562
+ ++G+ I+WM +M F M KK+ C NL L+IL
Sbjct: 121 ILNVAQQLEVGQSINWMFKMHFSMGIFKKKFICINLLVLRIL 162
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 90.9 bits (224), Expect = 1e-17
Identities = 51/153 (33%), Positives = 86/153 (55%), Gaps = 5/153 (3%)
Query: 160 VNSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALE 219
+++ ++ C GI + L+ P+T NG SER IR+ RT++ A L SFW A+
Sbjct: 556 LSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVL 615
Query: 220 MATYLMNILPHKTI--NFESPLKVLYHKDPSYNHLRVFGCLCFPLFPSSKIYKLQPRSTK 277
ATYL+N +P + + + ++P ++ ++K P HLRVFG + + +K K +S K
Sbjct: 616 TATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRVFGATVY-VHIKNKQGKFDDKSFK 674
Query: 278 CVFLGYPSNHRGYKCLGLLSNKIIICRHVLFNE 310
+F+GY N G+K ++ K I+ R V+ +E
Sbjct: 675 SIFVGYEPN--GFKLWDAVNEKFIVARDVVVDE 705
Score = 60.5 bits (145), Expect = 2e-08
Identities = 30/74 (40%), Positives = 43/74 (57%)
Query: 440 NWKMAMNDEYNSLIKNKTWDLVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLVGDGAG 499
+W+ A+N E N+ N TW + RP + N++ S W+F K G R+KARLV G
Sbjct: 905 SWEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFT 964
Query: 500 QQVGIDCGETFSPM 513
Q+ ID ETF+P+
Sbjct: 965 QKYQIDYEETFAPV 978
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 52.0 bits (123), Expect = 8e-06
Identities = 47/174 (27%), Positives = 77/174 (44%), Gaps = 9/174 (5%)
Query: 132 QYLKN*EHTSTHNFKEKLKQFNVTMAVN-VNSNFQKMCEANGIVFRLSCPYTSSQNGKSE 190
Q KN ++ T F K+++ N N ++ + GI L+ + NG++E
Sbjct: 674 QVRKNIQYVETQ-FDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAE 732
Query: 191 RKIRSTNNIIRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLKVLYHKDPSYN 250
R IR+ TLL ++L FW +A+ AT + N L HK+ + PLK + + +
Sbjct: 733 RYIRTIITDATTLLRQSNLRVKFWEYAVTSATNIRNYLEHKSTG-KLPLKAISRQPVT-- 789
Query: 251 HLRVFGCLCF---PLFPSSKIYKLQPRSTKCVFLGYPSNHRGYKCLGLLSNKII 301
+R+ L F + + KL+P + L N GYK NKI+
Sbjct: 790 -VRLMSFLPFGEKGIIWNHNHKKLKPSGLPSIILCKDPNSYGYKFFIPSKNKIV 842
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 49.7 bits (117), Expect = 4e-05
Identities = 26/78 (33%), Positives = 42/78 (53%), Gaps = 3/78 (3%)
Query: 200 IRTLLIHASLPSSFWHHALEMATYLMNILPHKTINFESPLKVLYHKDPSYNHLRVFGCLC 259
+R++L LP +F A A +++N P INF P +V + P+Y++LR FGC+
Sbjct: 9 VRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYLRRFGCVA 68
Query: 260 FPLFPSSKIYKLQPRSTK 277
+ KL+PR+ K
Sbjct: 69 YIHCDEG---KLKPRAKK 83
>YAIA_SCHPO (Q09897) Hypothetical protein C24B11.10c in chromosome I
Length = 932
Score = 42.4 bits (98), Expect = 0.006
Identities = 37/125 (29%), Positives = 53/125 (41%), Gaps = 12/125 (9%)
Query: 346 QTGP--PIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNL--PPIFQPTLQANTKP 401
QT P PIP P H + + S P+++SP ++PI NL P F P Q + K
Sbjct: 432 QTSPLSPIP-PVHTTHGLVSDIRPLPSVSSPIMRADSTPISHNLAVTPSFSPIPQQSNKS 490
Query: 402 ITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEYNSLIKNKTWDLV 461
I + N + + V S RN +S + + N E + K KT D +
Sbjct: 491 IGNHK------NTSVANVSSKVANSKFERN-ISMMSNSNASSPAIFEVGAEAKEKTKDAI 543
Query: 462 PRPPD 466
P PD
Sbjct: 544 PCHPD 548
>ATX2_HUMAN (Q99700) Ataxin-2 (Spinocerebellar ataxia type 2 protein)
Length = 1312
Score = 39.3 bits (90), Expect = 0.050
Identities = 35/150 (23%), Positives = 60/150 (39%), Gaps = 26/150 (17%)
Query: 314 PYAKLHIPQPNTYTFLDNELSPYIIQH-----LMDQTQTGPPIPQPAHQPNHITSPCPNS 368
P + + Q TY + N QH + + GPPI T P ++
Sbjct: 984 PMTPMPVNQAKTYRAVPNMPQQRQDQHHQSAMMHPASAAGPPIAA--------TPPAYST 1035
Query: 369 PNITSPPQSNPTSPIQQNL-------PPIFQPTLQANTK---PITRSQHGIFKPNQKYYG 418
+ PQ P P+ Q++ P ++ P +Q N + P T +Q G+ + YG
Sbjct: 1036 QYVAYSPQQFPNQPLVQHVPHYQSQHPHVYSPVIQGNARMMAPPTHAQPGLVSSSATQYG 1095
Query: 419 LH--THVTKSPLPRNPVSALKDPNWKMAMN 446
H TH + P+ P + P++ A++
Sbjct: 1096 AHEQTHAMYA-CPKLPYNKETSPSFYFAIS 1124
>PCLO_HUMAN (Q9Y6V0) Piccolo protein (Aczonin)
Length = 5183
Score = 37.4 bits (85), Expect = 0.19
Identities = 35/97 (36%), Positives = 39/97 (40%), Gaps = 19/97 (19%)
Query: 346 QTGPPIPQPA------HQPNHITSPCPNSPNITSPPQSNPTS--PIQQNLPPIFQPTLQA 397
QT PP QP QP P P P T PP P S P Q P +P+ Q
Sbjct: 442 QTKPPSQQPGSTKPPPQQPGP-AKPSPQQPGSTKPPSQQPGSAKPSAQQPSPA-KPSAQQ 499
Query: 398 NTKPITRSQHGIFKPNQKYYGLHTHVTKSPLPRNPVS 434
TKP+ SQ G KP Q T SP + P S
Sbjct: 500 FTKPV--SQTGFGKPLQP-------PTVSPSAKQPPS 527
Score = 34.3 bits (77), Expect = 1.6
Identities = 21/68 (30%), Positives = 30/68 (43%), Gaps = 4/68 (5%)
Query: 343 DQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNP--TSPIQQNLPPIFQPTLQANTK 400
+++Q GP +P QP+ +T P P PP P T P Q L P P Q ++
Sbjct: 322 EKSQPGPA--KPPAQPSGLTKPLAQQPGTVKPPVQPPGTTKPPAQPLGPAKPPAQQTGSE 379
Query: 401 PITRSQHG 408
+ Q G
Sbjct: 380 KPSSEQPG 387
>AUT2_HUMAN (Q8WXX7) Autism susceptibility gene 2 protein
Length = 1259
Score = 37.4 bits (85), Expect = 0.19
Identities = 29/108 (26%), Positives = 45/108 (40%), Gaps = 7/108 (6%)
Query: 303 CRHVLFNENIF----PYAKLHIPQPNTYTFLDNEL-SPYIIQHLMDQTQTGPPIPQPAHQ 357
C+ +F + P IPQP T L P ++Q Q P QP
Sbjct: 285 CKEPIFEPVVLKDPCPQVAQPIPQPQTEPQLRAPSPDPDLVQRTEAPPQPPPLSTQPPQG 344
Query: 358 PNHITSPCPNSPNITSPPQ-SNPTSPIQQNLPPI-FQPTLQANTKPIT 403
P P + PP+ +PT + QNLPP+ P+ Q+ ++P++
Sbjct: 345 PPEAQLQPAPQPQVQRPPRPQSPTQLLHQNLPPVQAHPSAQSLSQPLS 392
>POLG_PPVRA (P17767) Genome polyprotein [Contains: P1 proteinase
(N-terminal protein); Helper component proteinase (EC
3.4.22.45) (HC-pro); Protein P3; 6 kDa protein 1 (6K1);
Cytoplasmic inclusion protein (CI); 6 kDa protein 2
(6K2); Viral genome-linked p
Length = 3140
Score = 37.0 bits (84), Expect = 0.25
Identities = 25/96 (26%), Positives = 44/96 (45%), Gaps = 6/96 (6%)
Query: 337 IIQHLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQN-LPPIFQPTL 395
++ H D+ + + A +P+ +T+P SP + PP P + L PIF P
Sbjct: 2806 VVVHQADEREDEEEVD--AGKPSVVTAPAATSPILQPPPVIQPAPRTTASMLNPIFTP-- 2861
Query: 396 QANTKPITRSQHGIFKPNQKYYGLHTHVTKSPLPRN 431
A T+P T+ + +P + +G + + SP N
Sbjct: 2862 -ATTQPATKPVSQVSQPQLQTFGTYGNEDASPSNSN 2896
>EXTN_SORBI (P24152) Extensin precursor (Proline-rich glycoprotein)
Length = 283
Score = 37.0 bits (84), Expect = 0.25
Identities = 42/152 (27%), Positives = 54/152 (34%), Gaps = 30/152 (19%)
Query: 317 KLHIPQPNTYTFLDNELSPYIIQHLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQ 376
K H P P TYT P P P+P P + SP P SP PP+
Sbjct: 65 KEHKPTPPTYTPSPKPTPP-------------PATPKPT-PPTYTPSPKPKSPVYPPPPK 110
Query: 377 -SNPTSPIQQNLPPIFQPTLQANTKPITRS--QHGIFKPNQKYYGLHTHVTKSPLPRNPV 433
S P + PP +P KP ++ P+ K VTK P P+ P
Sbjct: 111 ASTPPTYTPSPKPPATKPPTYPTPKPPATKPPTPPVYTPSPK-----PPVTKPPTPK-PT 164
Query: 434 SALKDPNWKMAMNDEYNSLIKNKTWDLVPRPP 465
+ PN K + K T P+PP
Sbjct: 165 PPVYTPNPKPPVT-------KPPTHTPSPKPP 189
Score = 33.9 bits (76), Expect = 2.1
Identities = 28/87 (32%), Positives = 35/87 (40%), Gaps = 18/87 (20%)
Query: 349 PPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQ--QNLPPIFQP-TLQANTKPITRS 405
PP +P P + SP P +T PP PT P+ PP+ +P T + KP T
Sbjct: 136 PPATKPPTPPVYTPSP---KPPVTKPPTPKPTPPVYTPNPKPPVTKPPTHTPSPKPPTS- 191
Query: 406 QHGIFKPNQKYYGLHTHVTKSPLPRNP 432
KP Y T SP P P
Sbjct: 192 -----KPTPPVY------TPSPKPPKP 207
Score = 33.5 bits (75), Expect = 2.8
Identities = 30/102 (29%), Positives = 43/102 (41%), Gaps = 17/102 (16%)
Query: 347 TGPPIPQPA---HQPNH---ITSPCPNSPNITSPPQSNPTSPI-------QQNLPPIFQP 393
T PP P+P + PN +T P ++P+ PP S PT P+ + PP + P
Sbjct: 156 TKPPTPKPTPPVYTPNPKPPVTKPPTHTPS-PKPPTSKPTPPVYTPSPKPPKPSPPTYTP 214
Query: 394 TLQ--ANTKPITRSQHGIFKPNQKYYGLHTHVTK-SPLPRNP 432
T + A P + H P+ Y H K +P P P
Sbjct: 215 TPKPPATKPPTSTPTHPKPTPHTPYPQAHPPTYKPAPKPSPP 256
>YNM4_YEAST (P53919) Hypothetical 54.9 kDa protein in SPC98-TOM70
intergenic region
Length = 492
Score = 36.2 bits (82), Expect = 0.43
Identities = 23/68 (33%), Positives = 29/68 (41%), Gaps = 8/68 (11%)
Query: 311 NIFPYAKLHIPQPNTYTFLDNELSPYIIQHLMDQTQTGPPIPQPAHQPNHITSPCPNSPN 370
N PY L P Y N +SP Q M TQ+ PP+P N + +P P
Sbjct: 400 NTVPYGSL----PPAY----NNMSPPTQQSFMPMTQSQPPLPYGVPPMNQMQNPMYIQPP 451
Query: 371 ITSPPQSN 378
+PPQ N
Sbjct: 452 PQAPPQGN 459
>CAP2_DICDI (P34122) CABP1-related protein p31/p34
Length = 287
Score = 36.2 bits (82), Expect = 0.43
Identities = 44/189 (23%), Positives = 71/189 (37%), Gaps = 28/189 (14%)
Query: 353 QPAHQPNHITSPCPNSPNITSPPQSNPTSP---IQQNLPPIFQPTLQANTKPITRSQHGI 409
QP+ P +++P P + PP SN +P + PP P Q + P + Q
Sbjct: 19 QPSSTPG-VSNPNPQANQFLPPPPSNTQAPRPGFPPSAPPPSAPAGQYSMPPPPQQQQ-- 75
Query: 410 FKPNQKYYGLHTHVTKSPLPRNPVSALKDPNWKMAMNDEY-NSLIKNKTWDLVPRPP--- 465
YG+ + S + VS +KD ++ D Y L WD+ P
Sbjct: 76 ---QAGQYGMPPPPSGSGIGTG-VSLVKDQQISLSKEDPYLRKLTVGLGWDVNTTPSAPF 131
Query: 466 DVNVIRSMW-------------IFRHKEKSDGTFERHKARLVGDGAGQQVGIDCG-ETFS 511
D++ + M + +K+ DG+ H L G G G + + S
Sbjct: 132 DLDAVVFMLGANGMVRQPADFIFYNNKQSRDGSIFHHGDNLTGAGDGDDEVVSVNLQAVS 191
Query: 512 PM*NQLLFA 520
P +L+FA
Sbjct: 192 PDVTRLVFA 200
>SMR1_MOUSE (Q61900) Submaxillary gland androgen regulated protein 1
precursor (Salivary protein MSG1)
Length = 147
Score = 35.8 bits (81), Expect = 0.56
Identities = 23/55 (41%), Positives = 28/55 (50%), Gaps = 7/55 (12%)
Query: 348 GPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQP-TLQANTKP 401
GP I +P P PCP P PP SNP+ P ++PP P T+QA T P
Sbjct: 67 GPGIGRPPPPP-----PCPPVPPHPRPP-SNPSPPPTPSIPPTGPPTTVQATTMP 115
>P32_MYCGE (Q49417) P32 adhesin (Cytadhesin P32)
Length = 280
Score = 35.8 bits (81), Expect = 0.56
Identities = 25/83 (30%), Positives = 36/83 (43%), Gaps = 9/83 (10%)
Query: 335 PYIIQHLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLP-PIFQP 393
P + Q L Q Q P+ +PA PN P N PN P +N + N+ P F P
Sbjct: 142 PQVQQPLQPQFQQRVPLLRPAFNPNMQQRPGFNQPNQQFQPHNNFNPRMNPNMQRPGFNP 201
Query: 394 TLQANTKPITRSQHGIFKPNQKY 416
+Q + G +PNQ++
Sbjct: 202 NMQ--------QRPGFNQPNQQF 216
>CYAA_USTMA (P49606) Adenylate cyclase (EC 4.6.1.1) (ATP
pyrophosphate-lyase) (Adenylyl cyclase)
Length = 2493
Score = 35.8 bits (81), Expect = 0.56
Identities = 27/99 (27%), Positives = 42/99 (42%), Gaps = 9/99 (9%)
Query: 350 PIPQPAHQPNHITSPCPNSPNITSPPQSNPTSPIQQNLPPIFQPTL-QANTKPITRS--- 405
P+ PA NH T NSP+ S P N PP P + +A++ P S
Sbjct: 338 PMRPPAWLDNHCTLANSNSPSSASLRSHYHQPPASSNPPPWQNPLVSRADSTPSAMSLED 397
Query: 406 ----QHGIFK-PNQKYYGLHTHVTKSPLPRNPVSALKDP 439
+H + K P ++ G+ H+ K+ P + +DP
Sbjct: 398 EVEAEHHLKKDPRKRIKGVRHHLAKTTKPGEDADSARDP 436
>ADF1_CANAL (P46589) Adherence factor (Adhesion and aggregation
mediating surface antigen)
Length = 612
Score = 35.8 bits (81), Expect = 0.56
Identities = 34/95 (35%), Positives = 43/95 (44%), Gaps = 17/95 (17%)
Query: 321 PQPNTYTFLDNELSPYIIQHLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPT 380
PQPN F DN + Y+I M+QT P Q QPN + N ++ PQ
Sbjct: 135 PQPNNMQFFDNTIPNYLI---MNQT-ISPSQTQTTAQPN------ISYYNYSTQPQLQSA 184
Query: 381 SPIQQNLPPIFQPTLQANTKP---ITRSQHGIFKP 412
PI + P QP QA T+P +RSQ KP
Sbjct: 185 QPISHSQP---QPQPQA-TQPRSNRSRSQTSFSKP 215
>YO61_CAEEL (P34600) Hypothetical protein ZK1098.1 in chromosome III
Length = 724
Score = 35.4 bits (80), Expect = 0.73
Identities = 38/154 (24%), Positives = 64/154 (40%), Gaps = 23/154 (14%)
Query: 323 PNTYTFLDNEL--SPYIIQHLMDQTQTGPPIPQPAHQPNHITSPCPNSPNITSPPQSNPT 380
PN ++FL+ L + I Q L++Q PP+ A P + P + ++ +P + PT
Sbjct: 3 PNGFSFLNPNLVAAANIQQVLLNQRFGMPPVGSIAQVPL-LQMP---THSVVAPHVAAPT 58
Query: 381 SPIQQNLPPIFQPTLQANTKPITRSQHGIFKPNQKYYGLHTHVTK-----------SPLP 429
P +PP ++++ P S + + H VTK +PL
Sbjct: 59 RPSPMLVPPGMGID-ESHSSPSVESDWSVHTNEKGTPYYHNRVTKQTSWIKPDVLKTPLE 117
Query: 430 RNPVSALKDPNWKMAMNDE-----YNSLIKNKTW 458
R+ + WK M+D+ YN+L K W
Sbjct: 118 RSTSGQPQQGQWKEFMSDDGKPYYYNTLTKKTQW 151
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 35.4 bits (80), Expect = 0.73
Identities = 22/68 (32%), Positives = 32/68 (46%)
Query: 161 NSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEM 220
N K E NGI + S +G +ER R+ + RT L + LP+ W A+E
Sbjct: 316 NRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF 375
Query: 221 ATYLMNIL 228
+T + N L
Sbjct: 376 STIVRNSL 383
Score = 33.5 bits (75), Expect = 2.8
Identities = 27/60 (45%), Positives = 30/60 (50%), Gaps = 12/60 (20%)
Query: 444 AMNDEYNSLIKNKTWDLVPRPPD------VNVIRSMWIFRHKEKSDGTFERHKARLVGDG 497
A + E N L+K KTWD R D VI SM+IF K DGT HKAR V G
Sbjct: 826 AYHKEVNQLLKMKTWD-TDRYYDRKEIDPKRVINSMFIFNRKR--DGT---HKARFVARG 879
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 35.4 bits (80), Expect = 0.73
Identities = 22/68 (32%), Positives = 32/68 (46%)
Query: 161 NSNFQKMCEANGIVFRLSCPYTSSQNGKSERKIRSTNNIIRTLLIHASLPSSFWHHALEM 220
N K E NGI + S +G +ER R+ + RT L + LP+ W A+E
Sbjct: 316 NRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEF 375
Query: 221 ATYLMNIL 228
+T + N L
Sbjct: 376 STIVRNSL 383
Score = 33.5 bits (75), Expect = 2.8
Identities = 26/63 (41%), Positives = 31/63 (48%), Gaps = 18/63 (28%)
Query: 444 AMNDEYNSLIKNKTWD---------LVPRPPDVNVIRSMWIFRHKEKSDGTFERHKARLV 494
A + E N L+K KTWD + P+ VI SM+IF K DGT HKAR V
Sbjct: 826 AYHKEVNQLLKMKTWDTDKYYDRKEIDPK----RVINSMFIFNRKR--DGT---HKARFV 876
Query: 495 GDG 497
G
Sbjct: 877 ARG 879
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.337 0.147 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,679,526
Number of Sequences: 164201
Number of extensions: 4650826
Number of successful extensions: 19238
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 87
Number of HSP's that attempted gapping in prelim test: 18479
Number of HSP's gapped (non-prelim): 499
length of query: 952
length of database: 59,974,054
effective HSP length: 120
effective length of query: 832
effective length of database: 40,269,934
effective search space: 33504585088
effective search space used: 33504585088
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC146853.2


