

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146842.6 - phase: 0
(535 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
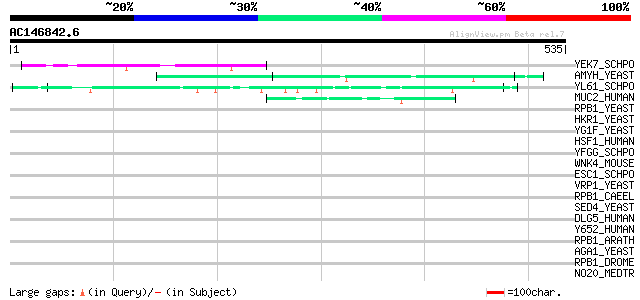
Score E
Sequences producing significant alignments: (bits) Value
YEK7_SCHPO (O36019) Protein C4F10.07c in chromosome I 61 8e-09
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 50 1e-05
YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein P... 47 1e-04
MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2) 46 2e-04
RPB1_YEAST (P04050) DNA-directed RNA polymerase II largest subun... 44 0.001
HKR1_YEAST (P41809) Hansenula MRAKII killer toxin-resistant prot... 44 0.001
YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A... 43 0.002
HSF1_HUMAN (Q00613) Heat shock factor protein 1 (HSF 1) (Heat sh... 43 0.002
YFGG_SCHPO (O13854) Hypothetical serine/threonine-rich protein C... 43 0.002
WNK4_MOUSE (Q80UE6) Serine/threonine-protein kinase WNK4 (EC 2.7... 43 0.002
ESC1_SCHPO (Q04635) Protein esc1 42 0.004
VRP1_YEAST (P37370) Verprolin 42 0.005
RPB1_CAEEL (P16356) DNA-directed RNA polymerase II largest subun... 42 0.005
SED4_YEAST (P25365) SED4 protein 41 0.007
DLG5_HUMAN (Q8TDM6) Discs, large homolog 5 (Placenta and prostat... 41 0.009
Y652_HUMAN (O75143) Hypothetical protein KIAA0652 40 0.015
RPB1_ARATH (P18616) DNA-directed RNA polymerase II largest subun... 40 0.015
AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor 40 0.015
RPB1_DROME (P04052) DNA-directed RNA polymerase II largest subun... 40 0.020
NO20_MEDTR (P93329) Early nodulin 20 precursor (N-20) 39 0.034
>YEK7_SCHPO (O36019) Protein C4F10.07c in chromosome I
Length = 758
Score = 60.8 bits (146), Expect = 8e-09
Identities = 63/252 (25%), Positives = 106/252 (42%), Gaps = 44/252 (17%)
Query: 12 AAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLAL 71
+AK Q+I F K+ II+ESR N S P +S ++ KWFNL +
Sbjct: 37 SAKLGQVIHHCFYKTGLIILESRL------NVFGTSRPRESSKNN--------KWFNLEI 82
Query: 72 RECPTALENTDLWRHSNLQPI-VVDVVLVHRNLSFSPKVRS---FVKERNPFEEFGGGSE 127
E E +W++ L P + +++H L S ++ V + +
Sbjct: 83 VETELYAEQFKIWKNIELSPSRKIPPMVLHTYLDISDLSKNQTLSVSDGTHSHAINFNNM 142
Query: 128 QNEKIV-ERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLL 186
KIV ERW++ + + + S+ L LYKK +L RSLY L+
Sbjct: 143 STMKIVLERWIVNLDGEAL--------------STPLELAVLYKKLVVLFRSLYTYTHLM 188
Query: 187 PAFKIFKELSS-SANVSAFSLAHRVST----------FFEPFTTKQEAEMLKFVFTPVDT 235
P +K+ ++ A+ ++ + +ST P ++ + + F F+PV T
Sbjct: 189 PLWKLKSKIHKLRAHGTSLKVGCALSTDDVLSNDFLPISAPISSSLGSSIATFSFSPVGT 248
Query: 236 NSGKLCLSVMYR 247
+G +SV YR
Sbjct: 249 PAGDFRISVQYR 260
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 50.4 bits (119), Expect = 1e-05
Identities = 59/238 (24%), Positives = 88/238 (36%), Gaps = 12/238 (5%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SS P T+ +P+T S PV S + SS P S + E+
Sbjct: 606 SSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSS 665
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSL-LHKKNVNFDEYYPS 372
A P S S T S S + S+ + P +S S + + P
Sbjct: 666 APVPTP---SSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPV 722
Query: 373 PNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIR 432
P S S + SSS+P+ S + S SAPV E + +P T + S+
Sbjct: 723 PTPSSSTTESSSAPV----PTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAP 778
Query: 433 ISRCTSETERSRN----LMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSS 486
+ +S T S + S TT + + S + AP+S+P S + SSS
Sbjct: 779 VPTPSSSTTESSSAPVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSS 836
Score = 47.8 bits (112), Expect = 7e-05
Identities = 80/377 (21%), Positives = 129/377 (33%), Gaps = 16/377 (4%)
Query: 142 SKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIFKELSSSANV 201
S + S+ T++TT SS + + S+ + S P + S++
Sbjct: 412 SSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSA 471
Query: 202 SAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPL 261
S S+ P + E T T S + S+ TP
Sbjct: 472 PVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPAPTPS 531
Query: 262 SPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNY 321
S T+ +P+T S PV S + SS P + + S +P +
Sbjct: 532 SST--TESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTSSTTESSSAPVPTPSSSTT 589
Query: 322 YS---PSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPS 378
S P PT S S T S+A + PSS S + + P+P+ S +
Sbjct: 590 ESSSAPVPTPSSSTTESSSAPAP---TPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTT 646
Query: 379 PSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTS 438
S S+ P S + S SAPV P++ T S + S+ ++ T+
Sbjct: 647 ESSSAPVP------TPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTT 700
Query: 439 ETERSR-NLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDTD 497
E+ + S TT + S + AP +P S + SSS ++
Sbjct: 701 ESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESS 760
Query: 498 FTCPFDVDDDDTTDPGS 514
+ P TT+ S
Sbjct: 761 -SAPVPTPSSSTTESSS 776
Score = 42.4 bits (98), Expect = 0.003
Identities = 81/379 (21%), Positives = 122/379 (31%), Gaps = 55/379 (14%)
Query: 149 SSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAH 208
SS+ T++T+ S+ T + S+ T K ++S + H
Sbjct: 230 SSTTTSSTSESSTTTSSTSESSTSSSTTAPATPTTTSCTKEKPTPPTTTSCTKEKPTPPH 289
Query: 209 RVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITD 268
+T T K TPV T S + SS P T+
Sbjct: 290 HDTTPCTKKKTTTSKTCTKKTTTPVPTPSSS-----------TTESSSAPVPTPSSSTTE 338
Query: 269 YVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTH 328
+P+T S PV S + SS P + + S S + +P PT
Sbjct: 339 SSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTP 398
Query: 329 SESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIY 388
S S T S+A SS P S S + SSS+P+
Sbjct: 399 SSSTTESSSAPVTSSTTESSSAPVTS-----------------------STTESSSAPVT 435
Query: 389 SLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSE--------- 439
S + S SAPV E + +P T + S+ ++ T+E
Sbjct: 436 S-------STTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTP 488
Query: 440 ----TERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDD 495
TE S + S TT + S + AP +P S + SSS +
Sbjct: 489 SSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTE 548
Query: 496 TDFTCPFDVDDDDTTDPGS 514
+ + P TT+ S
Sbjct: 549 SS-SAPVPTPSSSTTESSS 566
Score = 40.0 bits (92), Expect = 0.015
Identities = 62/220 (28%), Positives = 82/220 (37%), Gaps = 37/220 (16%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
SS P T+ +P+T S PV S + SS P S + E+
Sbjct: 732 SSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSS 791
Query: 314 ASPPAVNY------YSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFD 367
A P + +P PT S S + S+A PSS P S S
Sbjct: 792 APVPTPSSSTTESSVAPVPTPSSSSNITSSA-------PSSTPFSSSTES---------- 834
Query: 368 EYYPSPNFSPSPSPSSSSPIYSL---GSLA-----SKTLLRSESAPVNIPNAEVT--YSP 417
P P S S + SSS+P+ S S+A S + + SAP +IP + T +S
Sbjct: 835 SSVPVPTPSSSTTESSSAPVSSSTTESSVAPVPTPSSSSNITSSAPSSIPFSSTTESFST 894
Query: 418 GHT---SRQNLPPS-TPIRISRCTSETERSRNLMQSCTTP 453
G T S P S T +S T T S TTP
Sbjct: 895 GTTVTPSSSKYPGSQTETSVSSTTETTIVPTKTTTSVTTP 934
>YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein
PB15E9.01c in chromosome I precursor
Length = 943
Score = 47.0 bits (110), Expect = 1e-04
Identities = 113/504 (22%), Positives = 190/504 (37%), Gaps = 57/504 (11%)
Query: 3 TTSSHAQTEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRP 62
TTS + T +A + + + A S + S +L++SS N +T +SP+S+S +SSS+
Sbjct: 60 TTSYNYNTSSASSSSLTSSSAASSS--LTSSSSLASSSTNSTTSASPTSSSLTSSSAT-- 115
Query: 63 RDKWFNLALRECPTALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEF 122
++ L S + + +L+ S S + +
Sbjct: 116 -----------------SSSLASSSTTSSSLASSSITSSSLASSSITSSSLASSSTTSSS 158
Query: 123 GGGSEQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYAT 182
S N S + ++SN+A ++ +S++ N +T SL +T
Sbjct: 159 LASSSTNSTTSATPTSSATSSSLSSTAASNSATSSSLASSSL--NSTTSATATSSSLSST 216
Query: 183 VRLLPAFKIFKELSS-----SANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNS 237
A SS SA ++ S++ VS+ P T+ T T++
Sbjct: 217 AASNSATSSSLASSSLNSTTSATATSSSISSTVSSS-TPLTSSNS--------TTAATSA 267
Query: 238 GKLCLSVMYRPCVSDVSSEPT-TPLSPQ---VITDYVGSPLTD----RLMRFPSLPVVRM 289
S Y SS P+ TPLS T +PLT S P+ +
Sbjct: 268 SATSSSAQYNTSSLLPSSTPSSTPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSV 327
Query: 290 PSHGS----SPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCP 345
S S S SS P S +S + + +S P + S + T S S T +++ +S
Sbjct: 328 SSANSTTATSTSSTPLSSVNSTTATSA-SSTPLTSVNSTTAT-SASSTPLTSVNSTSATS 385
Query: 346 PSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAP 405
SS P + + + + PS N S S P+SS L S S T + S P
Sbjct: 386 ASSTPLTSANSTT----STSVSSTAPSYNTS-SVLPTSSVSSTPLSSANSTTATSASSTP 440
Query: 406 VNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQK 465
++ N+ S T ++ +T S + S + +TP + +
Sbjct: 441 LSSVNSTTATSASSTPLSSVNSTTATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSA 500
Query: 466 YSGGKIAPNSSPQISFSRSSSRSY 489
S + NS+ S S S++ SY
Sbjct: 501 SSTPLTSANSTTSTSVS-STAPSY 523
Score = 45.1 bits (105), Expect = 5e-04
Identities = 100/478 (20%), Positives = 175/478 (35%), Gaps = 48/478 (10%)
Query: 37 SASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLALRECPT-------ALENTDLWRHSNL 89
+ SS + S+L+S S+ SSS +SS N PT + ++ L S
Sbjct: 66 NTSSASSSSLTSSSAASSSLTSSSSLASSSTNSTTSASPTSSSLTSSSATSSSLASSSTT 125
Query: 90 QPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCS 149
+ + +L+ S S + + S N S + +
Sbjct: 126 SSSLASSSITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTSATPTSSATSSSLSSTA 185
Query: 150 SSNTANTTRRSSNTFLQNLYKKSTLLLRSL-------YATVRLLPAFKIFKELSSSANVS 202
+SN+A ++ +S++ N +T SL AT L + + S++A S
Sbjct: 186 ASNSATSSSLASSSL--NSTTSATATSSSLSSTAASNSATSSSLASSSLNSTTSATATSS 243
Query: 203 AFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLC---------LSVMYRPCVSDV 253
+ S ST + A + N+ L LS +
Sbjct: 244 SISSTVSSSTPLTSSNSTTAATSASATSSSAQYNTSSLLPSSTPSSTPLSSANSTTATSA 303
Query: 254 SSEPTTPLSPQVITDYVGSPLTD-------RLMRFPSLPVVRMPS-HGSSPSSLPFSRRH 305
SS P T ++ T +PL+ S P+ + S +S SS P + +
Sbjct: 304 SSTPLTSVNSTTTTSASSTPLSSVSSANSTTATSTSSTPLSSVNSTTATSASSTPLTSVN 363
Query: 306 SWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVN 365
S + + +S P + S S T S S T +++A+S SS P + S+L +V+
Sbjct: 364 STTATSA-SSTPLTSVNSTSAT-SASSTPLTSANSTTSTSVSSTAPSYNTSSVLPTSSVS 421
Query: 366 FDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNL 425
+P S + + ++S+ L S+ S T + S P++ N+ S T ++
Sbjct: 422 -----STPLSSANSTTATSASSTPLSSVNSTTATSASSTPLSSVNSTTATSASSTPLTSV 476
Query: 426 -------PPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSS 476
STP+ TS T S + S + S+ + Y+ + P SS
Sbjct: 477 NSTTATSASSTPLTSVNSTSATSASSTPLTSANSTTST-SVSSTAPSYNTSSVLPTSS 533
>MUC2_HUMAN (Q02817) Mucin 2 precursor (Intestinal mucin 2)
Length = 5179
Score = 46.2 bits (108), Expect = 2e-04
Identities = 47/186 (25%), Positives = 73/186 (38%), Gaps = 27/186 (14%)
Query: 248 PCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSW 307
P ++ + PTT SP T P T PS P + +S ++LP + S
Sbjct: 1589 PTITTTTPPPTTTPSPPTTTTTTPPPTTT-----PSPPTTTPITPPTSTTTLPPTTTPS- 1642
Query: 308 SYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFD 367
+PP SP T + S + + + PP++ P P +
Sbjct: 1643 PPPTTTTTPPPTTTPSPPTTTTPSPPITTTTTP----PPTTTPSSPITTT---------- 1688
Query: 368 EYYPSPNFS----PSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQ 423
PSP + PSP+ + SSPI + + +S T + P+ T SP T+
Sbjct: 1689 ---PSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPPPTTMTTPSPTTTPSPPTTTMT 1745
Query: 424 NLPPST 429
LPP+T
Sbjct: 1746 TLPPTT 1751
Score = 40.0 bits (92), Expect = 0.015
Identities = 49/208 (23%), Positives = 80/208 (37%), Gaps = 35/208 (16%)
Query: 231 TPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDY--VGSPLTDRLMRFPSLPVVR 288
TP+ T + +S P + S PTT SP T + T PS P+
Sbjct: 1447 TPLPTTTPSPPISTTTTPPPTTTPSPPTTTPSPPTTTPSPPTTTTTTPPPTTTPSPPMTT 1506
Query: 289 MPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSES---HTLVSNASSQGYCP 345
+ +S ++LP SPP +P PT + S T ++ +S P
Sbjct: 1507 PITPPASTTTLP---------PTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTSTTTLP 1557
Query: 346 PSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAP 405
P++ P P + P P +PSP P++++P + T+ + P
Sbjct: 1558 PTTTPSPPPTTTTT-----------PPPTTTPSP-PTTTTP-------SPPTITTTTPPP 1598
Query: 406 VNIPN--AEVTYSPGHTSRQNLPPSTPI 431
P+ T +P T+ + P +TPI
Sbjct: 1599 TTTPSPPTTTTTTPPPTTTPSPPTTTPI 1626
Score = 39.7 bits (91), Expect = 0.020
Identities = 39/146 (26%), Positives = 61/146 (41%), Gaps = 21/146 (14%)
Query: 323 SPSPTHSESHTLVSNASSQGYCPP-------SSLPPHPSEMSLLHKKNVNFDEYYPSPNF 375
+P+PT +++ T ++ P + P H S + N P P
Sbjct: 4164 TPTPTGTQTPTTTPITTTTTVTPTPTPTGTQTGPPTHTSTAPIAELTTSN-----PPPES 4218
Query: 376 S-PSPSPSSSSPIYSLGSLASK---TLLRSESAPVNIPNAEVTYSPGHT-----SRQNLP 426
S P S S+SSP+ +L S + + +AP + P A T S GHT S P
Sbjct: 4219 STPQTSRSTSSPLTESTTLLSTLPPAIEMTSTAPPSTPTAPTTTSGGHTLSPPPSTTTSP 4278
Query: 427 PSTPIRISRCTSETERSRNLMQSCTT 452
P TP R + S + + + +Q+ TT
Sbjct: 4279 PGTPTRGTTTGSSSAPTPSTVQTTTT 4304
Score = 37.0 bits (84), Expect = 0.13
Identities = 50/245 (20%), Positives = 83/245 (33%), Gaps = 28/245 (11%)
Query: 231 TPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRL----------MR 280
TP T + ++ P S + PTT SP T P T
Sbjct: 1493 TPPPTTTPSPPMTTPITPPASTTTLPPTTTPSPPTTTTTTPPPTTTPSPPTTTPITPPTS 1552
Query: 281 FPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
+LP PS + ++ P SPP + +P PT + S + +
Sbjct: 1553 TTTLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPSPPTITTTTPPPTTTPSPPTTTTTTP 1612
Query: 341 QGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKT--- 397
PP++ P P+ + + P+ SP P+ +++ P + S + T
Sbjct: 1613 ----PPTTTPSPPTTTPITPPTSTT--TLPPTTTPSPPPTTTTTPPPTTTPSPPTTTTPS 1666
Query: 398 ---LLRSESAPVNIPNAEVTYSPG------HTSRQNLPPSTPIRISRCTSETERSRNLMQ 448
+ P P++ +T +P T PS+PI + S T
Sbjct: 1667 PPITTTTTPPPTTTPSSPITTTPSPPTTTMTTPSPTTTPSSPITTTTTPSSTTTPSPPPT 1726
Query: 449 SCTTP 453
+ TTP
Sbjct: 1727 TMTTP 1731
>RPB1_YEAST (P04050) DNA-directed RNA polymerase II largest subunit
(EC 2.7.7.6) (RNA polymerase II subunit 1) (B220)
Length = 1733
Score = 43.5 bits (101), Expect = 0.001
Identities = 49/146 (33%), Positives = 65/146 (43%), Gaps = 10/146 (6%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRH-SWSYENC 312
S PT+P Y SP + P+ P S SP+S +S S+S +
Sbjct: 1576 SYSPTSPSYSPTSPSY--SPTSPSYS--PTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSP 1631
Query: 313 RASPPAVNYYSPSPTHSESHTLVSNASSQGYCP--PSSLPPHPSEMSLLHKKNVNFDEYY 370
SP + +Y SP++S + S +S Y P PS P PS + Y
Sbjct: 1632 SYSPTSPSYSPTSPSYSPTSPSYS-PTSPAYSPTSPSYSPTSPSYSPTSPSYSPTSPSYS 1690
Query: 371 P-SPNFSP-SPSPSSSSPIYSLGSLA 394
P SPN+SP SPS S +SP YS GS A
Sbjct: 1691 PTSPNYSPTSPSYSPTSPGYSPGSPA 1716
>HKR1_YEAST (P41809) Hansenula MRAKII killer toxin-resistant protein
1 precursor
Length = 1802
Score = 43.5 bits (101), Expect = 0.001
Identities = 114/513 (22%), Positives = 189/513 (36%), Gaps = 92/513 (17%)
Query: 10 TEAAKTEQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNL 69
+E+ +T+ T F S+ E A+S S + S SS + S S++ DK +
Sbjct: 229 SESTQTDFSNTVSFENSVE---EEYAMSKSQLSESYSSSSTVYSGGESTA----DKTSSS 281
Query: 70 ALRECPTALENTDLWRHSNLQPIVVDVVL---VHRNLSFSPKVRSFVKERNPFEEFGGGS 126
+ ++ T S + V V + + S S V+ ++ G
Sbjct: 282 PITSFSSSYSQTTSTETSESSRVAVGVSRPSSITQTTSIDSFSMSEVELSTYYDLSAGNY 341
Query: 127 EQNEKIVERWVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLL 186
E IV+R ++ +SS + R SNTF + + ++ S TV
Sbjct: 342 PDQELIVDRPATSSTAE-----TSSEASQGVSRESNTFAVSSISTTNFIVSSASDTV--- 393
Query: 187 PAFKIFKELSSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMY 246
+SS N +S H STF T+S S +Y
Sbjct: 394 -------VSTSSTNTVPYSSVH--STFVHA------------------TSSSTYISSSLY 426
Query: 247 RPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHS 306
S P+ LS V + + +P + F S+PV ++ SSPS+ S
Sbjct: 427 --------SSPS--LSASVSSHFGVAPFPSAYISFSSVPVAVSSTYTSSPSA---SVVVP 473
Query: 307 WSYENCRASPPAV-NYYSPSPTH----SESHTLVSNASSQGYCPPSSLPPHPSEMSLLHK 361
+Y + + P AV + Y+ SP+ S ++T +A +S P P+ +S +
Sbjct: 474 SAYASSPSVPVAVSSTYTSSPSAPAAISSTYTSSPSAPVAVSSTYTSSPSAPAAISSTYT 533
Query: 362 KNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLA----SKTLLRSESAPVNI-------PN 410
+ + S S +P++ S Y+ A S T S SAPV I P+
Sbjct: 534 SSPSAPVAVSSTYTSSPSAPAAISSTYTSSPSAPVAVSSTYTSSPSAPVAISSTYTSSPS 593
Query: 411 AEVTYSPGHTSRQNLP----------PSTPIRISRCTSETERSRNLMQSCTTPEKMFSIG 460
V S +TS + P PS P+ +S + + + + S T +
Sbjct: 594 VPVAVSSTYTSSPSAPAAISSTYTSSPSAPVAVSSTYTSSPSAPAAISSTYTSSPSVPVA 653
Query: 461 KESQKYSGGKIAP-------NSSPQISFSRSSS 486
S Y+ AP SSP + + SS+
Sbjct: 654 -VSSTYTSSPSAPAAISSTYTSSPSVPVAVSST 685
Score = 40.8 bits (94), Expect = 0.009
Identities = 51/179 (28%), Positives = 82/179 (45%), Gaps = 30/179 (16%)
Query: 251 SDVSSEPTTPLSPQVITDYVGSP-----LTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRH 305
S +S P+ P++ V + Y SP ++ PS PV ++ SSPS+
Sbjct: 698 STYTSSPSAPVA--VSSTYTSSPSAPAAISSTYTSSPSAPVAVSSTYTSSPSA---PAAI 752
Query: 306 SWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVN 365
S +Y + ++P AV S T++ S + + SS P + PS + +
Sbjct: 753 SSTYTSSPSAPVAV-----SSTYTSSPSALVVLSSTSTSSPYDIVYSPSTFAAISSG--- 804
Query: 366 FDEYYPSPNFSPS-PSPSSSSP---IYSLGSLASKTLLR----SESAPVNIPNAEVTYS 416
Y PSP+ S + S SSSSP +YSL S AS++ + S S ++P + TY+
Sbjct: 805 ---YTPSPSASVAMSSTSSSSPYDIVYSLSSSASRSSIATYEFSPSPSTSLPTSS-TYT 859
>YG1F_YEAST (P53214) Hypothetical 57.5 kDa protein in VMA7-RPS25A
intergenic region
Length = 551
Score = 43.1 bits (100), Expect = 0.002
Identities = 66/255 (25%), Positives = 105/255 (40%), Gaps = 27/255 (10%)
Query: 149 SSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAH 208
+SS+++ T SS++ + ST++ S LP F + S+S+ V++ +L+
Sbjct: 115 TSSSSSEVTSSSSSSSISPSSSSSTIISSS-----SSLPTFTV---ASTSSTVASSTLST 166
Query: 209 R----VSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQ 264
+ST FT E+ + + + T+ + S +Y P S +S P + S
Sbjct: 167 SSSLVISTSSSTFTFSSESSS-SLISSSISTS---VSTSSVYVP--SSSTSSPPSSSSEL 220
Query: 265 VITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSP 324
+ Y S + L + S S SS SS S S SY S + Y S
Sbjct: 221 TSSSYSSSSSSSTLFSYSSSFSSSSSSSSSSSSSSSSSSSSSSSYFTLSTSSSSSIYSSS 280
Query: 325 S-PTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDE-------YYPSPNFS 376
S P+ S S + +S SS+ P SE S L K + E YY + +S
Sbjct: 281 SYPSFSSSSSSNPTSSITSTSASSSITP-ASEYSNLAKTITSIIEGQTILSNYYTTITYS 339
Query: 377 PSPSPSSSSPIYSLG 391
P+ S SS + G
Sbjct: 340 PTASASSGKNSHHSG 354
Score = 38.1 bits (87), Expect = 0.058
Identities = 63/280 (22%), Positives = 110/280 (38%), Gaps = 16/280 (5%)
Query: 147 DCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIFKEL--SSSANVSAF 204
D +SS + +T +S +F S L L S ++ + ++ SSS+ V++
Sbjct: 66 DATSSASLSTPSIASVSFTSFPQSSSLLTLSSTLSSELSSSSMQVSSSSTSSSSSEVTSS 125
Query: 205 SLAHRVSTFFEPFTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQ 264
S + +S T + L FT T+S ++ + +S T S +
Sbjct: 126 SSSSSISPSSSSSTIISSSSSLP-TFTVASTSSTVASSTLSTSSSLVISTSSSTFTFSSE 184
Query: 265 VITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSP 324
+ + S ++ + S V +PS +S S S SY + +S +Y S
Sbjct: 185 SSSSLISSSISTSV----STSSVYVPSSSTSSPPSSSSELTSSSYSSSSSSSTLFSYSSS 240
Query: 325 SPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSS 384
+ S S + S++SS SS +S ++ YPS FS S SSS
Sbjct: 241 FSSSSSSSSSSSSSSSSSSSSSSSY----FTLSTSSSSSIYSSSSYPS--FSSS---SSS 291
Query: 385 SPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQN 424
+P S+ S ++ + + S N+ + G T N
Sbjct: 292 NPTSSITSTSASSSITPASEYSNLAKTITSIIEGQTILSN 331
Score = 31.2 bits (69), Expect = 7.1
Identities = 61/257 (23%), Positives = 92/257 (35%), Gaps = 24/257 (9%)
Query: 248 PCVSDVSSEPTTPLSPQVITDYVGSP--LTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRH 305
P S SS + S + + S + + PS+ V S S S L S
Sbjct: 40 PANSTTSSTAPSITSLSAVESFTSSTDATSSASLSTPSIASVSFTSFPQSSSLLTLSSTL 99
Query: 306 SWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNV- 364
S + S S T S S + S++SS P SS S S L V
Sbjct: 100 SSELSSSSMQ------VSSSSTSSSSSEVTSSSSSSSISPSSSSSTIISSSSSLPTFTVA 153
Query: 365 NFDEYYPSPNFSPSPSP--SSSSPIYSLGSLASKTLLRSE------SAPVNIPNAEVTYS 416
+ S S S S S+SS ++ S +S +L+ S ++ V +P++ +
Sbjct: 154 STSSTVASSTLSTSSSLVISTSSSTFTFSSESSSSLISSSISTSVSTSSVYVPSSSTSSP 213
Query: 417 PGHTSR-------QNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGG 469
P +S + ST S S + S + S ++ S + S
Sbjct: 214 PSSSSELTSSSYSSSSSSSTLFSYSSSFSSSSSSSSSSSSSSSSSSSSSSSYFTLSTSSS 273
Query: 470 KIAPNSSPQISFSRSSS 486
+SS SFS SSS
Sbjct: 274 SSIYSSSSYPSFSSSSS 290
>HSF1_HUMAN (Q00613) Heat shock factor protein 1 (HSF 1) (Heat shock
transcription factor 1) (HSTF 1)
Length = 529
Score = 43.1 bits (100), Expect = 0.002
Identities = 49/160 (30%), Positives = 66/160 (40%), Gaps = 33/160 (20%)
Query: 293 GSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNA-------------- 338
GS+ S +SR+ +S E+ S P Y +PSP +S S +A
Sbjct: 217 GSAHSMPKYSRQ--FSLEHVHGSGP---YSAPSPAYSSSSLYAPDAVASSGPIISDITEL 271
Query: 339 --SSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSP-----SPS-PSSSSPIYSL 390
+S P S+ P S L V E PSP SP SP PSS + S
Sbjct: 272 APASPMASPGGSIDERPLSSSPL----VRVKEEPPSPPQSPRVEEASPGRPSSVDTLLSP 327
Query: 391 GSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTP 430
+L L SE AP ++ +T + GHT + PPS P
Sbjct: 328 TALIDSILRESEPAPASV--TALTDARGHTDTEGRPPSPP 365
>YFGG_SCHPO (O13854) Hypothetical serine/threonine-rich protein
C19G12.16c in chromosome I precursor
Length = 670
Score = 42.7 bits (99), Expect = 0.002
Identities = 101/466 (21%), Positives = 180/466 (37%), Gaps = 29/466 (6%)
Query: 37 SASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLALRECPTALENTDLWRHSNLQPIVVDV 96
++S+ +FST+++ S SSSS + ++N + + + + L S L ++
Sbjct: 193 ASSTSSFSTITNTSMIPSSSSFTTTTGSPYYNTSSFLPSSVISSASLSSSSVLPTSIITS 252
Query: 97 VLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQNEKIVERWVIQYESKKIKDCSSSNTANT 156
+S S + SF + G + Y + + S ++ ++
Sbjct: 253 TSTPVTVS-SSSLSSFTPSYSTNLTTTGSTTTTGSATVSSSPFYSNSSVIPTSVPSSVSS 311
Query: 157 TRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIFKELSSSANVSAFSLAHRVSTFFEP 216
SS+++ L +T + + T F S+++V S+ VS+F
Sbjct: 312 FTSSSSSYTTTLTASNTSVTYTGTGTGSA--TFTSSPPFYSNSSVIPTSVPSSVSSFTSS 369
Query: 217 ---FTTKQEAEMLKFVFTPVDTNSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSP 273
+TT A +T T S S P S+ S PT+ P ++ + S
Sbjct: 370 NSSYTTTLTASNTSITYTGTGTGSATFTSSP---PFYSNSSVIPTSV--PSSVSSFTSSN 424
Query: 274 LTDRLMRFPSLPVVRMPSHGSSP----SSLPFSRRHSWSYENCRASPPAVNYYSPS---- 325
+ S V G+ SS PF +S S ++P +V+ ++ S
Sbjct: 425 SSYTTTLTASNTTVTFTGTGTGSATFTSSPPF---YSNSSVIPTSAPSSVSSFTSSNSSY 481
Query: 326 -PTHSESHTLVS-NASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSS 383
T + S+T V+ + G +S P+ S S++ V+ S FS SPSP+S
Sbjct: 482 TTTLTASNTTVTFTGTGTGSATATSSSPYYSNSSIIVPTTVSTSGSVSS--FSSSPSPTS 539
Query: 384 S-SPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETER 442
S S +L S +++ + E T + T+ + P T +S TS
Sbjct: 540 SFSGTSALSSSSNEETTTTTQVTYTTSPEETTTTMTTTTCSSRPEETISTVS-TTSTVSE 598
Query: 443 SRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRS 488
S + S T+ ++ + S + +SS S SRSSS S
Sbjct: 599 SGSSSASITSTYPSSTLSMTTSHLSSSSV-HSSSAHSSSSRSSSMS 643
>WNK4_MOUSE (Q80UE6) Serine/threonine-protein kinase WNK4 (EC
2.7.1.37) (Protein kinase with no lysine 4) (Protein
kinase, lysine-deficient 4)
Length = 1222
Score = 42.7 bits (99), Expect = 0.002
Identities = 70/247 (28%), Positives = 98/247 (39%), Gaps = 62/247 (25%)
Query: 255 SEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRA 314
S P TPLSP SP T + P P+ +SPS P C
Sbjct: 797 SSPGTPLSPGAPF----SPGTPPVFPCPIFPI-------TSPSCYP-----------CPF 834
Query: 315 SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLP---PHPSEMSLLHKKNVNFDEYYP 371
S + N Y +P+ S +S+++SQ P SSLP P P F YP
Sbjct: 835 SQVSSNPYPQAPS---SLLPLSSSASQVPLPSSSLPISAPLP------------FSPSYP 879
Query: 372 SPNFSPSPSPSSSSPIYSLGSLASKTLLRSES----APVNIPNAEVTYSPGHTSRQNLP- 426
SP+ P SP SL S + LL S A + + + ++ SPG S Q+ P
Sbjct: 880 QDPLSPTSLPVCPSP-PSLPSTTAAPLLSLASAFSLAVMTVAQSLLSPSPGLLS-QSPPA 937
Query: 427 -----PSTPIRISRC--------TSETER--SRNLMQSCTTPEKMFSIGKESQKYSGGKI 471
PS P+ ++ C T+ETE SRN Q ++ I +E + G+
Sbjct: 938 PPGPLPSLPLSLASCDQESLSAQTAETENEASRNPAQPLLGDARLAPISEEGKPQLVGRF 997
Query: 472 APNSSPQ 478
SS +
Sbjct: 998 QVTSSKE 1004
>ESC1_SCHPO (Q04635) Protein esc1
Length = 413
Score = 42.0 bits (97), Expect = 0.004
Identities = 67/291 (23%), Positives = 112/291 (38%), Gaps = 40/291 (13%)
Query: 235 TNSGKLCLSVMYRPCVSDVS-SEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHG 293
T + + L M +P S S S +TP SPQ + PLT S P+ S
Sbjct: 12 TPTSSIPLRQMSQPTTSAPSNSASSTPYSPQQV------PLTHN-----SYPLSTPSSFQ 60
Query: 294 SSPSSLP--------FSRRHSW---SYENCRASPPAVNYYSPS-PTHSESHTLVSNASSQ 341
+ LP F+R W S +SP + + + P H+ + + ++SS
Sbjct: 61 HGQTRLPPINCLAEPFNRPQPWHSNSAAPASSSPTSATLSTAAHPVHTNAAQVAGSSSSY 120
Query: 342 GYCPPSSLPPH---PSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTL 398
Y S+PP S+ S H + + S +PS + SSS+ + S S+++
Sbjct: 121 VY----SVPPTNSTTSQASAKHSAVPHRSSQFQSTTLTPSTTDSSSTDVSSSDSVSTSAS 176
Query: 399 LRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETE-----RSRNLMQSCTTP 453
+ S V++ + S T N P +S+ + T + + QS P
Sbjct: 177 SSNASNTVSVTSPA---SSSATPLPNQPSQQQFLVSKNDAFTTFVHSVHNTPMQQSMYVP 233
Query: 454 EKMFSIGKESQKYSGGKIAPNSSP-QISFSRSSSRSYQDEFDDTDFTCPFD 503
++ S + + P SP Q S+S+ SY + + P D
Sbjct: 234 QQQTSHSSGASYQNESANPPVQSPMQYSYSQGQPFSYPQHKNQSFSASPID 284
>VRP1_YEAST (P37370) Verprolin
Length = 817
Score = 41.6 bits (96), Expect = 0.005
Identities = 60/252 (23%), Positives = 91/252 (35%), Gaps = 40/252 (15%)
Query: 257 PTTPLSPQVITDYVGS----PLTD-------RLMRFPSLPVVRMPSHGSSPSSLPFSRRH 305
P PLSP + S P+ D + PS P +P G+S +P +R H
Sbjct: 136 PNAPLSPAPAVPSIPSSSAPPIPDIPSSAAPPIPIVPSSPAPPLPLSGASAPKVPQNRPH 195
Query: 306 ---------SWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEM 356
S ++ S P+V+ +P + T VSN PP + PP P+
Sbjct: 196 MPSVRPAHRSHQRKSSNISLPSVS--APPLPSASLPTHVSN-------PPQAPPPPPTPT 246
Query: 357 SLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYS 416
L KN+ + SP PSS P L LA RSE V ++ +
Sbjct: 247 IGLDSKNIKPTDNAVSP-------PSSEVPAGGLPFLAEINARRSERGAVEGVSSTKIQT 299
Query: 417 PGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSS 476
H S P P+ S T + L + P + ++ +K + P
Sbjct: 300 ENHKS----PSQPPLPSSAPPIPTSHAPPLPPTAPPPPSLPNVTSAPKKATSAPAPPPPP 355
Query: 477 PQISFSRSSSRS 488
+ S +S+ S
Sbjct: 356 LPAAMSSASTNS 367
>RPB1_CAEEL (P16356) DNA-directed RNA polymerase II largest subunit
(EC 2.7.7.6)
Length = 1852
Score = 41.6 bits (96), Expect = 0.005
Identities = 55/192 (28%), Positives = 77/192 (39%), Gaps = 21/192 (10%)
Query: 243 SVMYRPCVSDVSSEPTTP-LSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPF 301
S Y P S S P++P SP T SP P+ P S SP+S +
Sbjct: 1658 SPSYSP--SSPSYSPSSPRYSPTSPTYSPTSPTYS-----PTSPTYSPTSPTYSPTSPSY 1710
Query: 302 SRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPS-SLPPHPSEMSLLH 360
+S + + SP + Y SP++S + S S Q Y P S + P +
Sbjct: 1711 ESGGGYSPSSPKYSPSSPTYSPTSPSYSPTSPQYSPTSPQ-YSPSSPTYTPSSPTYNPTS 1769
Query: 361 KKNVNFDEYYP-SPNFSP-SPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPG 418
+ + +Y P SP +SP SPS + SSP Y S T +P P YSP
Sbjct: 1770 PRGFSSPQYSPTSPTYSPTSPSYTPSSPQY------SPTSPTYTPSPSEQPGTSAQYSP- 1822
Query: 419 HTSRQNLPPSTP 430
+ PS+P
Sbjct: 1823 --TSPTYSPSSP 1832
Score = 41.2 bits (95), Expect = 0.007
Identities = 73/249 (29%), Positives = 96/249 (38%), Gaps = 51/249 (20%)
Query: 288 RMPSHGS------SPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQ 341
R PS+G SPSS FS SP +YSP+ S S++ S A+ Q
Sbjct: 1563 RTPSYGGMSPGVYSPSSPQFSM----------TSP----HYSPT---SPSYSPTSPAAGQ 1605
Query: 342 GYCPPSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYSLGSLASKTLL 399
PS P PS + Y P SP++SP SPS S +SP Y
Sbjct: 1606 SPVSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSYSPTSPSY----------- 1654
Query: 400 RSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSI 459
S S+P P++ +YSP S P++P TS T + S T+P +
Sbjct: 1655 -SPSSPSYSPSSP-SYSP---SSPRYSPTSPTYSP--TSPTYSPTSPTYSPTSP----TY 1703
Query: 460 GKESQKY-SGGKIAPNS---SPQISFSRSSSRSYQDEFDDTDFTCPFDVDDDDTTDPGSR 515
S Y SGG +P+S SP +S SY T P T P S
Sbjct: 1704 SPTSPSYESGGGYSPSSPKYSPSSPTYSPTSPSYSPTSPQYSPTSPQYSPSSPTYTPSSP 1763
Query: 516 YFITKIHRG 524
+ RG
Sbjct: 1764 TYNPTSPRG 1772
>SED4_YEAST (P25365) SED4 protein
Length = 1065
Score = 41.2 bits (95), Expect = 0.007
Identities = 73/349 (20%), Positives = 129/349 (36%), Gaps = 40/349 (11%)
Query: 192 FKELSSSANV-SAFSLAHRVSTFFEPF-------TTKQEAEML--------KFVFTPVDT 235
++ ++ SA++ SA +A + T F F TT+ E + + +F + T
Sbjct: 405 YRGITESADIISATDVASDIETEFSSFDTSTMRTTTEDEQKFVWISSSADSQFTSADIPT 464
Query: 236 NSGKLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPV----VRMPS 291
++ S V++EP IT + SP ++ PSLP+ + + S
Sbjct: 465 SASSSSSSSSSSFYEESVTNEPIVSSPTSEITKPLASPTEPNIVEKPSLPLNSESIDLLS 524
Query: 292 HGSSP---------------SSLPFSRRHSWSYENCRASPPAVNYYSPSPTH--SESHTL 334
S+ SSL + S + + ++ SPS +H S S +
Sbjct: 525 SSSNSITEYPEPTPDLEEKLSSLIVEQSESEITTDRESVSKLLSTESPSLSHMPSSSSSS 584
Query: 335 VSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFSPSPSPSSSSPIYSLGSLA 394
+S +SS P ++L + + N D + +F + P+S+ IY +
Sbjct: 585 LSLSSSLTTSPTTALSTSTATAVTTTQTNPTNDA--ANTSFLDNSKPASTREIYKTKIIT 642
Query: 395 SK-TLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTP 453
T + + P + NAE +S L P+ + S + + L + TP
Sbjct: 643 EVITKIEYRNIPASDSNAEAEQYVTTSSSMLLTPTDTMVSSPVSEIDPIASELERMVETP 702
Query: 454 EKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDTDFTCPF 502
SI E + I S S+ S S+ F D+ T F
Sbjct: 703 THSISIASEFDSVASNLIPNEEILSTSASQDSISSHPSTFSDSSITSGF 751
>DLG5_HUMAN (Q8TDM6) Discs, large homolog 5 (Placenta and prostate
DLG) (Discs large protein P-dlg)
Length = 1809
Score = 40.8 bits (94), Expect = 0.009
Identities = 65/232 (28%), Positives = 85/232 (36%), Gaps = 57/232 (24%)
Query: 242 LSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPF 301
LSV +P +P T PQ P D L+ P PS + P + P
Sbjct: 851 LSVYKKPKQRKSIFDPNTFKRPQT------PPKIDYLLPGPGPAHSPQPSKRAGPLTPPK 904
Query: 302 SRRHSWSYENCRASPPAVNYYSPSPTHSESH-TLVSNASSQGYCPPSSLPP-----HPSE 355
R S S + + T SES TLV ++ S PPS+LPP P
Sbjct: 905 PPRRSDS----------IKFQHRLETSSESEATLVGSSPSTS--PPSALPPDVDPGEPMH 952
Query: 356 MSLLHKKNVNF-DEYYPSPNFSPSPSPS--------SSSPIYSLGSLA----SKTLLRSE 402
S K V YYP + S P+ +S + LG S R +
Sbjct: 953 ASPPRKARVRIASSYYPEGDGDSSHLPAKKSCDEDLTSQKVDELGQKRRRPKSAPSFRPK 1012
Query: 403 SAPVNIP------------NAEVT--------YSPGHTSRQNLPPSTPIRIS 434
APV IP + E++ YSPGH+SR + PP P R S
Sbjct: 1013 LAPVVIPAQFLEEQKCVPASGELSPELQEWAPYSPGHSSRHSNPPLYPSRPS 1064
>Y652_HUMAN (O75143) Hypothetical protein KIAA0652
Length = 517
Score = 40.0 bits (92), Expect = 0.015
Identities = 56/262 (21%), Positives = 104/262 (39%), Gaps = 60/262 (22%)
Query: 16 EQIITEFFAKSLHIIIESRALSASSRNFSTLSSPSSTSSSSSSSVRPRDKWFNLALRECP 75
++ I F K++ +I+++R T SS S T S WFNLA+++ P
Sbjct: 14 DKFIKFFALKTVQVIVQARL----GEKICTRSSSSPTGSD----------WFNLAIKDIP 59
Query: 76 TALENTDLWRHSNLQPIVVDVVLVHRNLSFSPKVRSFVKERNPFEEFGGGSEQNEKIVER 135
H + + + V R++ +++ SE + +E
Sbjct: 60 EVT-------HEAKKALAGQLPAVGRSMCVEISLKT--------------SEGDSMELEI 98
Query: 136 WVIQYESKKIKDCSSSNTANTTRRSSNTFLQNLYKKSTLLLRSLYATVRLLPAFKIFKEL 195
W ++ K K+ S T +Y + +LLL+SL A R+ PA+++ ++
Sbjct: 99 WCLEMNEKCDKEIKVSYT--------------VYNRLSLLLKSLLAITRVTPAYRLSRKQ 144
Query: 196 SSSANVSAFSLAHRVSTFFEPFTTKQEAEMLKFV-FTPVDTNSGKLCLSVMYRPCVSDVS 254
+ + +R+ +F E + V V T G + LS YR ++ +S
Sbjct: 145 GHE-----YVILYRI--YFGEVQLSGLGEGFQTVRVGTVGTPVGTITLSCAYRINLAFMS 197
Query: 255 S---EPTTPLSPQVITDYVGSP 273
+ E T P+ +I +V P
Sbjct: 198 TRQFERTPPIMGIIIDHFVDRP 219
>RPB1_ARATH (P18616) DNA-directed RNA polymerase II largest subunit
(EC 2.7.7.6)
Length = 1840
Score = 40.0 bits (92), Expect = 0.015
Identities = 49/166 (29%), Positives = 66/166 (39%), Gaps = 19/166 (11%)
Query: 254 SSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENCR 313
S PT+P Y SP + P+ P S G SP+S +S
Sbjct: 1670 SYSPTSPSYSPTSPSY--SPTSPSYS--PTSPAYSPTSPGYSPTSPSYSPTSPSYGPTSP 1725
Query: 314 ASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYP-S 372
+ P YSPS +S S+ +S AS P+ P PS + Y P S
Sbjct: 1726 SYNPQSAKYSPSIAYSPSNARLSPASPYSPTSPNYSPTSPS-------YSPTSPSYSPSS 1778
Query: 373 PNFSP-SPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSP 417
P +SP SP S +SP YS + S TL P P++ Y+P
Sbjct: 1779 PTYSPSSPYSSGASPDYSPSAGYSPTL------PGYSPSSTGQYTP 1818
Score = 38.1 bits (87), Expect = 0.058
Identities = 62/211 (29%), Positives = 90/211 (42%), Gaps = 30/211 (14%)
Query: 282 PSLPVVRMPSHGSSPSSLPFS-RRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASS 340
P+ P S G SP+S +S +S + SP + +Y SP++S + S +S
Sbjct: 1561 PTSPGYSPTSPGYSPTSPTYSPSSPGYSPTSPAYSPTSPSYSPTSPSYSPTSPSYS-PTS 1619
Query: 341 QGYCP--PSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSP-SPSPSSSSPIYSLGSLASK 396
Y P PS P PS + Y P SP +SP SPS S +SP YS S +
Sbjct: 1620 PSYSPTSPSYSPTSPSYSPTSPAYSPTSPAYSPTSPAYSPTSPSYSPTSPSYSPTSPS-- 1677
Query: 397 TLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKM 456
S ++P P + +YSP P +P TS + S T+P
Sbjct: 1678 ---YSPTSPSYSPTSP-SYSP------TSPAYSP------TSPGYSPTSPSYSPTSP--- 1718
Query: 457 FSIGKESQKYSGGKIAPNSSPQISFSRSSSR 487
S G S Y+ + SP I++S S++R
Sbjct: 1719 -SYGPTSPSYN--PQSAKYSPSIAYSPSNAR 1746
>AGA1_YEAST (P32323) A-agglutinin attachment subunit precursor
Length = 725
Score = 40.0 bits (92), Expect = 0.015
Identities = 44/180 (24%), Positives = 74/180 (40%), Gaps = 23/180 (12%)
Query: 317 PAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPSPNFS 376
P + S + + + + T +S+ S+ PSS PS S + + S
Sbjct: 168 PVTSTLSSTTSSNPTTTSLSSTSTS----PSSTSTSPSSTSTSSSSTSTSSSSTSTSSSS 223
Query: 377 PSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRC 436
S SPSS+S SL S +S + S+S+ + ++ + SP TS + ST S+
Sbjct: 224 TSTSPSSTSTSSSLTSTSSSSTSTSQSS-TSTSSSSTSTSPSSTSTSSSSTSTS-PSSKS 281
Query: 437 TSETERSRNLMQSCTTPEKMFSIGKESQKYSGGKIAPNSSPQISFSRSSSRSYQDEFDDT 496
TS + S + + T+P +SSP ++ + SS S F D+
Sbjct: 282 TSASSTSTSSYSTSTSPS-----------------LTSSSPTLASTSPSSTSISSTFTDS 324
Score = 32.0 bits (71), Expect = 4.1
Identities = 65/272 (23%), Positives = 103/272 (36%), Gaps = 28/272 (10%)
Query: 253 VSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSSLPFSRRHSWSYENC 312
VSS P S +I SP+T L S S+P++ S S S +
Sbjct: 153 VSSSAIEPSSASII-----SPVTSTLSSTTS----------SNPTTTSLSST-STSPSST 196
Query: 313 RASPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLPPHPSEMSLLHKKNVNFDEYYPS 372
SP + + S S + S S T S++S+ PSS S S +
Sbjct: 197 STSPSSTSTSSSSTSTSSSSTSTSSSSTS--TSPSSTSTSSSLTSTSSSSTSTSQSSTST 254
Query: 373 PNFSPSPSPSSSSPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPGHTSRQ-NLPPSTPI 431
+ S S SPSS+S S S + + S S+ + + + SP TS L ++P
Sbjct: 255 SSSSTSTSPSSTSTSSSSTSTSPSSKSTSASS-TSTSSYSTSTSPSLTSSSPTLASTSPS 313
Query: 432 RISRCTSETERSRNLMQSCTTPEKMFSIGKESQK-YSGGKIAPNSSPQISFSRSSSRSYQ 490
S ++ T+ + +L S + S+ S YS P++S ++ +S + +
Sbjct: 314 STSISSTFTDSTSSLGSSIASSSTSVSLYSPSTPVYS----VPSTSSNVATPSMTSSTVE 369
Query: 491 DEFDDTDFTCPFDVDDDDTTDPG---SRYFIT 519
+ TT P S YF T
Sbjct: 370 TTVSSQSSSEYITKSSISTTIPSFSMSTYFTT 401
>RPB1_DROME (P04052) DNA-directed RNA polymerase II largest subunit
(EC 2.7.7.6)
Length = 1887
Score = 39.7 bits (91), Expect = 0.020
Identities = 53/212 (25%), Positives = 84/212 (39%), Gaps = 23/212 (10%)
Query: 282 PSLPVVRMPSHGSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLVSNASSQ 341
P++ PS S S++ +S +++S + SP + +Y SP++S S S +S
Sbjct: 1647 PTVQFQSSPSFAGSGSNI-YSPGNAYSPSSSNYSPNSPSYSPTSPSYSPSSPSYS-PTSP 1704
Query: 342 GYCP--PSSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSPSPSPSSSSPIYSLGSLASKTL 398
Y P PS P P+ + Y P SPN+S SP S +SP YS +
Sbjct: 1705 CYSPTSPSYSPTSPNYTPVT-------PSYSPTSPNYSASPQYSPASPAYSQTGVKYSPT 1757
Query: 399 LRSESAPVNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERS-RNLMQSCTTPEKMF 457
+ S P SP + P TP + + S + + S ++P+
Sbjct: 1758 SPTYSPP----------SPSYDGSPGSPQYTPGSPQYSPASPKYSPTSPLYSPSSPQHSP 1807
Query: 458 SIGKESQKYSGGKIAPNSSPQISFSRSSSRSY 489
S + +P SP +S SS Y
Sbjct: 1808 SNQYSPTGSTYSATSPRYSPNMSIYSPSSTKY 1839
Score = 37.7 bits (86), Expect = 0.075
Identities = 57/201 (28%), Positives = 81/201 (39%), Gaps = 28/201 (13%)
Query: 293 GSSPSSLPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHT------LVSNASSQGYCPP 346
GS+PS P + NC Y+SP P H + T S AS P
Sbjct: 1497 GSTPSMTP----PMTPWANCNTP----RYFSP-PGHVSAMTPGGPSFSPSAASDASGMSP 1547
Query: 347 SSLPPHPSEMSLLHKKNVNFDEYYP-SPNFSPSPSPSSSSPIYSLGSLASKTLLRSESAP 405
S P HP S + Y+P SP+ SPS SP +SP Y+ S + S S+P
Sbjct: 1548 SWSPAHPG--SSPSSPGPSMSPYFPASPSVSPSYSP--TSPNYTASSPGGASPNYSPSSP 1603
Query: 406 VNIPNAEVTYSPGHTSRQNLPPSTPIRISRCTSETERSRNLMQSCTTPEKMFSIGKESQK 465
P + + SP + S +TP + T + S S T+P ++ +S
Sbjct: 1604 NYSPTSPLYASPRYAS------TTPNFNPQSTGYSPSSSG--YSPTSPVYSPTVQFQSSP 1655
Query: 466 YSGGKIAPNSSPQISFSRSSS 486
G + SP ++S SSS
Sbjct: 1656 SFAGSGSNIYSPGNAYSPSSS 1676
>NO20_MEDTR (P93329) Early nodulin 20 precursor (N-20)
Length = 268
Score = 38.9 bits (89), Expect = 0.034
Identities = 36/110 (32%), Positives = 48/110 (42%), Gaps = 8/110 (7%)
Query: 315 SPPAVNYYSPSPTHSESHTLVSNASSQGYCPPSSLP-PHPSEMSLLHKKNVNFDEYYPSP 373
SPP +P P H +L S S PS P P P + H + + PSP
Sbjct: 141 SPPTPRSSTPIP-HPPRRSLPSPPSPSPSPSPSPSPSPSPRSTPIPHPRKRSPASPSPSP 199
Query: 374 NFSPSPSPSSS-----SPIYSLGSLASKTLLRSESAPVNIPNAEVTYSPG 418
+ S SPSPS S SP S+ SLA + S+ +P P+ + S G
Sbjct: 200 SLSKSPSPSESPSLAPSPSDSVASLAPSS-SPSDESPSPAPSPSSSGSKG 248
Score = 36.2 bits (82), Expect = 0.22
Identities = 46/147 (31%), Positives = 57/147 (38%), Gaps = 25/147 (17%)
Query: 239 KLCLSVMYRPCVSDVSSEPTTPLSPQVITDYVGSPLTDRLMRFPSLPVVRMPSHGSSPSS 298
KL + VM P +S P+ P +P+ T P R + P P PS SPS
Sbjct: 122 KLAVVVMVAPVLSSPPPPPSPP-TPRSSTPIPHPPR--RSLPSPPSPSPS-PSPSPSPSP 177
Query: 299 LPFSRRHSWSYENCRASPPAVNYYSPSPTHSESHTLV-SNASSQGYCPPSSLPPHPSEMS 357
P S + ASP S SP+ SES +L S + S PSS P S
Sbjct: 178 SPRSTPIPHPRKRSPASPSPSPSLSKSPSPSESPSLAPSPSDSVASLAPSSSPSDESP-- 235
Query: 358 LLHKKNVNFDEYYPSPNFSPSPSPSSS 384
SP+PSPSSS
Sbjct: 236 ------------------SPAPSPSSS 244
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.127 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,224,083
Number of Sequences: 164201
Number of extensions: 2828819
Number of successful extensions: 10121
Number of sequences better than 10.0: 205
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 196
Number of HSP's that attempted gapping in prelim test: 9081
Number of HSP's gapped (non-prelim): 721
length of query: 535
length of database: 59,974,054
effective HSP length: 115
effective length of query: 420
effective length of database: 41,090,939
effective search space: 17258194380
effective search space used: 17258194380
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Medicago: description of AC146842.6


