

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146842.3 + phase: 0 /pseudo
(57 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
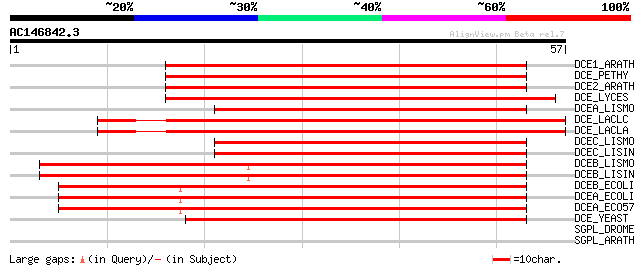
Score E
Sequences producing significant alignments: (bits) Value
DCE1_ARATH (Q42521) Glutamate decarboxylase 1 (EC 4.1.1.15) (GAD 1) 73 2e-13
DCE_PETHY (Q07346) Glutamate decarboxylase (EC 4.1.1.15) (GAD) 72 2e-13
DCE2_ARATH (Q42472) Glutamate decarboxylase 2 (EC 4.1.1.15) (GAD 2) 72 2e-13
DCE_LYCES (P54767) Glutamate decarboxylase (EC 4.1.1.15) (GAD) (... 71 5e-13
DCEA_LISMO (Q9F5P3) Glutamate decarboxylase alpha (EC 4.1.1.15) ... 61 6e-10
DCE_LACLC (O30418) Glutamate decarboxylase (EC 4.1.1.15) 59 2e-09
DCE_LACLA (Q9CG20) Glutamate decarboxylase (EC 4.1.1.15) (GAD) 59 2e-09
DCEC_LISMO (Q8Y4K4) Probabl glutamate decarboxylase gamma (EC 4.... 55 3e-08
DCEC_LISIN (Q928K4) Probabl glutamate decarboxylase gamma (EC 4.... 55 3e-08
DCEB_LISMO (Q9EYW9) Glutamate decarboxylase beta (EC 4.1.1.15) (... 55 3e-08
DCEB_LISIN (Q928R9) Glutamate decarboxylase beta (EC 4.1.1.15) (... 55 3e-08
DCEB_ECOLI (P28302) Glutamate decarboxylase beta (EC 4.1.1.15) (... 55 3e-08
DCEA_ECOLI (P80063) Glutamate decarboxylase alpha (EC 4.1.1.15) ... 55 3e-08
DCEA_ECO57 (P58228) Glutamate decarboxylase alpha (EC 4.1.1.15) ... 55 3e-08
DCE_YEAST (Q04792) Glutamate decarboxylase (EC 4.1.1.15) (GAD) 47 1e-05
SGPL_DROME (Q9V7Y2) Sphingosine-1-phosphate lyase (EC 4.1.2.27) ... 28 4.5
SGPL_ARATH (Q9C509) Sphingosine-1-phosphate lyase (EC 4.1.2.27) ... 27 7.7
>DCE1_ARATH (Q42521) Glutamate decarboxylase 1 (EC 4.1.1.15) (GAD 1)
Length = 502
Score = 72.8 bits (177), Expect = 2e-13
Identities = 32/37 (86%), Positives = 32/37 (86%)
Query: 17 FPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
F Y LEWDFRLPLVKSINVSGHKYGL Y GIGWVIW
Sbjct: 254 FLYPELEWDFRLPLVKSINVSGHKYGLVYAGIGWVIW 290
>DCE_PETHY (Q07346) Glutamate decarboxylase (EC 4.1.1.15) (GAD)
Length = 500
Score = 72.4 bits (176), Expect = 2e-13
Identities = 31/37 (83%), Positives = 32/37 (85%)
Query: 17 FPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
F Y LEWDFRLPLVKSINVSGHKYGL Y GIGWV+W
Sbjct: 254 FIYPELEWDFRLPLVKSINVSGHKYGLVYAGIGWVVW 290
>DCE2_ARATH (Q42472) Glutamate decarboxylase 2 (EC 4.1.1.15) (GAD 2)
Length = 494
Score = 72.4 bits (176), Expect = 2e-13
Identities = 31/37 (83%), Positives = 32/37 (85%)
Query: 17 FPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
F Y LEWDFRLPLVKSINVSGHKYGL Y GIGWV+W
Sbjct: 253 FIYPELEWDFRLPLVKSINVSGHKYGLVYAGIGWVVW 289
>DCE_LYCES (P54767) Glutamate decarboxylase (EC 4.1.1.15) (GAD) (ERT
D1)
Length = 502
Score = 71.2 bits (173), Expect = 5e-13
Identities = 31/40 (77%), Positives = 34/40 (84%)
Query: 17 FPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIWCSR 56
F + LEWDFRLPLVKSINVSGHKYGL Y G+GWVIW S+
Sbjct: 255 FLWPDLEWDFRLPLVKSINVSGHKYGLVYAGVGWVIWRSK 294
>DCEA_LISMO (Q9F5P3) Glutamate decarboxylase alpha (EC 4.1.1.15)
(GAD-alpha)
Length = 462
Score = 60.8 bits (146), Expect = 6e-10
Identities = 24/32 (75%), Positives = 27/32 (84%)
Query: 22 LEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
LEWDFRLP V SIN SGHKYGL Y G+GW++W
Sbjct: 255 LEWDFRLPNVVSINTSGHKYGLVYPGVGWILW 286
>DCE_LACLC (O30418) Glutamate decarboxylase (EC 4.1.1.15)
Length = 466
Score = 58.9 bits (141), Expect = 2e-09
Identities = 28/48 (58%), Positives = 33/48 (68%), Gaps = 3/48 (6%)
Query: 10 LHAPTSSFPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIWCSRR 57
L+AP F LEWDFRL V SIN SGHKYGL Y G+GWV+W ++
Sbjct: 250 LYAP---FVEPELEWDFRLKNVISINTSGHKYGLVYPGVGWVLWRDKK 294
>DCE_LACLA (Q9CG20) Glutamate decarboxylase (EC 4.1.1.15) (GAD)
Length = 466
Score = 58.9 bits (141), Expect = 2e-09
Identities = 28/48 (58%), Positives = 33/48 (68%), Gaps = 3/48 (6%)
Query: 10 LHAPTSSFPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIWCSRR 57
L+AP F LEWDFRL V SIN SGHKYGL Y G+GWV+W ++
Sbjct: 250 LYAP---FVEPELEWDFRLKNVISINTSGHKYGLVYPGVGWVLWRDKK 294
>DCEC_LISMO (Q8Y4K4) Probabl glutamate decarboxylase gamma (EC
4.1.1.15) (GAD-gamma)
Length = 467
Score = 55.5 bits (132), Expect = 3e-08
Identities = 22/32 (68%), Positives = 25/32 (77%)
Query: 22 LEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
L WDFRL V SIN SGHKYGL Y G+GW++W
Sbjct: 260 LPWDFRLKNVVSINTSGHKYGLVYPGVGWILW 291
>DCEC_LISIN (Q928K4) Probabl glutamate decarboxylase gamma (EC
4.1.1.15) (GAD-gamma)
Length = 467
Score = 55.5 bits (132), Expect = 3e-08
Identities = 22/32 (68%), Positives = 25/32 (77%)
Query: 22 LEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
L WDFRL V SIN SGHKYGL Y G+GW++W
Sbjct: 260 LPWDFRLKNVVSINTSGHKYGLVYPGVGWILW 291
>DCEB_LISMO (Q9EYW9) Glutamate decarboxylase beta (EC 4.1.1.15)
(GAD-beta)
Length = 464
Score = 55.5 bits (132), Expect = 3e-08
Identities = 28/54 (51%), Positives = 33/54 (60%), Gaps = 4/54 (7%)
Query: 4 YPVTMDLHAPTSSFPYKMLE----WDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
Y V + + A + F +E WDFRL V SIN SGHKYGL Y GIGWV+W
Sbjct: 235 YKVYIHVDAASGGFFTPFVEPDIIWDFRLKNVISINTSGHKYGLVYPGIGWVLW 288
>DCEB_LISIN (Q928R9) Glutamate decarboxylase beta (EC 4.1.1.15)
(GAD-beta)
Length = 464
Score = 55.5 bits (132), Expect = 3e-08
Identities = 28/54 (51%), Positives = 33/54 (60%), Gaps = 4/54 (7%)
Query: 4 YPVTMDLHAPTSSFPYKMLE----WDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
Y V + + A + F +E WDFRL V SIN SGHKYGL Y GIGWV+W
Sbjct: 235 YKVYIHVDAASGGFFTPFVEPDIIWDFRLKNVISINTSGHKYGLVYPGIGWVLW 288
>DCEB_ECOLI (P28302) Glutamate decarboxylase beta (EC 4.1.1.15)
(GAD-beta)
Length = 466
Score = 55.5 bits (132), Expect = 3e-08
Identities = 26/54 (48%), Positives = 34/54 (62%), Gaps = 6/54 (11%)
Query: 6 VTMDLHAPTSS------FPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
+ +D+H +S F + WDFRLP VKSI+ SGHK+GL +G GWVIW
Sbjct: 236 IDIDMHIDAASGGFLAPFVAPDIVWDFRLPRVKSISASGHKFGLAPLGCGWVIW 289
>DCEA_ECOLI (P80063) Glutamate decarboxylase alpha (EC 4.1.1.15)
(GAD-alpha)
Length = 466
Score = 55.5 bits (132), Expect = 3e-08
Identities = 26/54 (48%), Positives = 34/54 (62%), Gaps = 6/54 (11%)
Query: 6 VTMDLHAPTSS------FPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
+ +D+H +S F + WDFRLP VKSI+ SGHK+GL +G GWVIW
Sbjct: 236 IDIDMHIDAASGGFLAPFVAPDIVWDFRLPRVKSISASGHKFGLAPLGCGWVIW 289
>DCEA_ECO57 (P58228) Glutamate decarboxylase alpha (EC 4.1.1.15)
(GAD-alpha)
Length = 466
Score = 55.5 bits (132), Expect = 3e-08
Identities = 26/54 (48%), Positives = 34/54 (62%), Gaps = 6/54 (11%)
Query: 6 VTMDLHAPTSS------FPYKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
+ +D+H +S F + WDFRLP VKSI+ SGHK+GL +G GWVIW
Sbjct: 236 IDIDMHIDAASGGFLAPFVAPDIVWDFRLPRVKSISASGHKFGLAPLGCGWVIW 289
>DCE_YEAST (Q04792) Glutamate decarboxylase (EC 4.1.1.15) (GAD)
Length = 585
Score = 46.6 bits (109), Expect = 1e-05
Identities = 19/35 (54%), Positives = 23/35 (65%)
Query: 19 YKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
Y M W F P V S+N SGHK+GL G+GWV+W
Sbjct: 297 YGMERWGFNHPRVVSMNTSGHKFGLTTPGLGWVLW 331
>SGPL_DROME (Q9V7Y2) Sphingosine-1-phosphate lyase (EC 4.1.2.27)
(SP-lyase) (SPL) (Sphingosine-1-phosphate aldolase)
Length = 545
Score = 28.1 bits (61), Expect = 4.5
Identities = 17/63 (26%), Positives = 29/63 (45%), Gaps = 8/63 (12%)
Query: 3 KYPVTMDLHAPTSSFP--------YKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIWC 54
KY + + + A SF YK+ +DF + V SI+ HKYG G +++
Sbjct: 297 KYDIPVHVDACLGSFVVALVRNAGYKLRPFDFEVKGVTSISADTHKYGFAPKGSSVILYS 356
Query: 55 SRR 57
++
Sbjct: 357 DKK 359
>SGPL_ARATH (Q9C509) Sphingosine-1-phosphate lyase (EC 4.1.2.27)
(SP-lyase) (SPL) (Sphingosine-1-phosphate aldolase)
Length = 544
Score = 27.3 bits (59), Expect = 7.7
Identities = 14/35 (40%), Positives = 21/35 (60%)
Query: 19 YKMLEWDFRLPLVKSINVSGHKYGLFYIGIGWVIW 53
Y++ +DF + V SI+V HKYGL G V++
Sbjct: 328 YQIPPFDFSVQGVTSISVDVHKYGLAPKGTSTVLY 362
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.328 0.143 0.501
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,350,976
Number of Sequences: 164201
Number of extensions: 212171
Number of successful extensions: 632
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 615
Number of HSP's gapped (non-prelim): 17
length of query: 57
length of database: 59,974,054
effective HSP length: 33
effective length of query: 24
effective length of database: 54,555,421
effective search space: 1309330104
effective search space used: 1309330104
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC146842.3


