

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146807.7 - phase: 0 /pseudo
(97 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
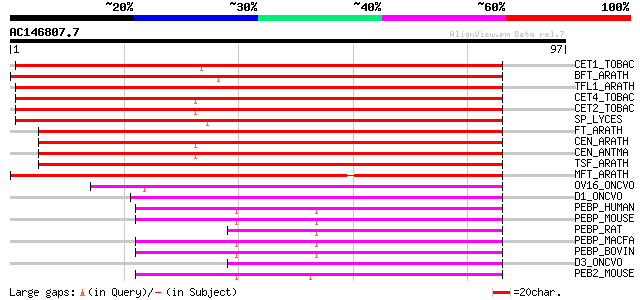
Score E
Sequences producing significant alignments: (bits) Value
CET1_TOBAC (Q9XH44) CEN-like protein 1 135 2e-32
BFT_ARATH (Q9FIT4) BROTHER of FT and TFL1 protein 122 1e-28
TFL1_ARATH (P93003) TERMINAL FLOWER 1 protein 119 2e-27
CET4_TOBAC (Q9XH42) CEN-like protein 4 118 3e-27
CET2_TOBAC (Q9XH43) CEN-like protein 2 117 5e-27
SP_LYCES (O82088) SELF-PRUNING protein 113 7e-26
FT_ARATH (Q9SXZ2) FLOWERING LOCUS T protein 112 1e-25
CEN_ARATH (Q9ZNV5) CENTRORADIALIS-like protein 111 4e-25
CEN_ANTMA (Q41261) CENTRORADIALIS protein 108 2e-24
TSF_ARATH (Q9S7R5) TWIN SISTER of FT protein (TFL1 like protein) 101 3e-22
MFT_ARATH (Q9XFK7) MOTHER of FT and TF1 protein 94 8e-20
OV16_ONCVO (P31729) OV-16 antigen precursor 52 3e-07
D1_ONCVO (P54186) D1 protein (Fragment) 52 3e-07
PEBP_HUMAN (P30086) Phosphatidylethanolamine-binding protein (PE... 47 8e-06
PEBP_MOUSE (P70296) Phosphatidylethanolamine-binding protein (PE... 47 1e-05
PEBP_RAT (P31044) Phosphatidylethanolamine-binding protein (PEBP... 46 1e-05
PEBP_MACFA (P48737) Phosphatidylethanolamine-binding protein (PE... 46 2e-05
PEBP_BOVIN (P13696) Phosphatidylethanolamine-binding protein (PE... 44 5e-05
D3_ONCVO (P54188) D3 protein (Fragment) 44 7e-05
PEB2_MOUSE (Q8VIN1) Phosphatidylethanolamine-binding protein-2 (... 44 9e-05
>CET1_TOBAC (Q9XH44) CEN-like protein 1
Length = 174
Score = 135 bits (340), Expect = 2e-32
Identities = 65/86 (75%), Positives = 75/86 (86%), Gaps = 1/86 (1%)
Query: 2 SRPLEPLSVGRVIGEVVDIFNPSVRMNVTYS-TKQVANGHELMPSIVMNKPRVDIGGEDM 60
SR +EPL V RVIGEVVD FNPSV++NV Y+ +KQV NGHELMP+++ KPRV+IGGEDM
Sbjct: 3 SRVVEPLVVARVIGEVVDSFNPSVKLNVIYNGSKQVFNGHELMPAVIAAKPRVEIGGEDM 62
Query: 61 RSAYTLIMTDPDAPSPSDPHLREHLH 86
RSAYTLIMTDPD P PSDP+LREHLH
Sbjct: 63 RSAYTLIMTDPDVPGPSDPYLREHLH 88
>BFT_ARATH (Q9FIT4) BROTHER of FT and TFL1 protein
Length = 177
Score = 122 bits (307), Expect = 1e-28
Identities = 56/87 (64%), Positives = 77/87 (88%), Gaps = 1/87 (1%)
Query: 1 MSRPLEPLSVGRVIGEVVDIFNPSVRMNVTYSTKQ-VANGHELMPSIVMNKPRVDIGGED 59
MSR +EPL VGRVIG+V+++FNPSV M VT+++ V+NGHEL PS++++KPRV+IGG+D
Sbjct: 1 MSREIEPLIVGRVIGDVLEMFNPSVTMRVTFNSNTIVSNGHELAPSLLLSKPRVEIGGQD 60
Query: 60 MRSAYTLIMTDPDAPSPSDPHLREHLH 86
+RS +TLIM DPDAPSPS+P++RE+LH
Sbjct: 61 LRSFFTLIMMDPDAPSPSNPYMREYLH 87
>TFL1_ARATH (P93003) TERMINAL FLOWER 1 protein
Length = 177
Score = 119 bits (297), Expect = 2e-27
Identities = 52/85 (61%), Positives = 69/85 (81%)
Query: 2 SRPLEPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMR 61
+R +EPL +GRV+G+V+D F P+ +MNV+Y+ KQV+NGHEL PS V +KPRV+I G D+R
Sbjct: 6 TRVIEPLIMGRVVGDVLDFFTPTTKMNVSYNKKQVSNGHELFPSSVSSKPRVEIHGGDLR 65
Query: 62 SAYTLIMTDPDAPSPSDPHLREHLH 86
S +TL+M DPD P PSDP L+EHLH
Sbjct: 66 SFFTLVMIDPDVPGPSDPFLKEHLH 90
>CET4_TOBAC (Q9XH42) CEN-like protein 4
Length = 175
Score = 118 bits (295), Expect = 3e-27
Identities = 57/86 (66%), Positives = 69/86 (79%), Gaps = 1/86 (1%)
Query: 2 SRPLEPLSVGRVIGEVVDIFNPSVRMNVTY-STKQVANGHELMPSIVMNKPRVDIGGEDM 60
S+ +PL +GRVIGEVVD F PSV+M+VTY S+K V NGHEL PS V +KPRV++ G D+
Sbjct: 3 SKMSDPLVIGRVIGEVVDYFTPSVKMSVTYNSSKHVYNGHELFPSSVTSKPRVEVHGGDL 62
Query: 61 RSAYTLIMTDPDAPSPSDPHLREHLH 86
RS +TLIM DPD P PSDP+LREHLH
Sbjct: 63 RSFFTLIMIDPDVPGPSDPYLREHLH 88
>CET2_TOBAC (Q9XH43) CEN-like protein 2
Length = 175
Score = 117 bits (293), Expect = 5e-27
Identities = 56/86 (65%), Positives = 69/86 (80%), Gaps = 1/86 (1%)
Query: 2 SRPLEPLSVGRVIGEVVDIFNPSVRMNVTY-STKQVANGHELMPSIVMNKPRVDIGGEDM 60
S+ +PL +GRVIGEVVD F PSV+M+VTY S+K V NGHEL PS V +KPRV++ G D+
Sbjct: 3 SKMSDPLVIGRVIGEVVDYFTPSVKMSVTYNSSKHVYNGHELFPSSVTSKPRVEVHGGDL 62
Query: 61 RSAYTLIMTDPDAPSPSDPHLREHLH 86
RS +T+IM DPD P PSDP+LREHLH
Sbjct: 63 RSFFTMIMIDPDVPGPSDPYLREHLH 88
>SP_LYCES (O82088) SELF-PRUNING protein
Length = 175
Score = 113 bits (283), Expect = 7e-26
Identities = 55/86 (63%), Positives = 66/86 (75%), Gaps = 1/86 (1%)
Query: 2 SRPLEPLSVGRVIGEVVDIFNPSVRMNVTYST-KQVANGHELMPSIVMNKPRVDIGGEDM 60
S+ EPL +GRVIGEVVD F PSV+M+V Y+ K V NGHE PS V +KPRV++ G D+
Sbjct: 3 SKMCEPLVIGRVIGEVVDYFCPSVKMSVVYNNNKHVYNGHEFFPSSVTSKPRVEVHGGDL 62
Query: 61 RSAYTLIMTDPDAPSPSDPHLREHLH 86
RS +TLIM DPD P PSDP+LREHLH
Sbjct: 63 RSFFTLIMIDPDVPGPSDPYLREHLH 88
>FT_ARATH (Q9SXZ2) FLOWERING LOCUS T protein
Length = 175
Score = 112 bits (281), Expect = 1e-25
Identities = 50/81 (61%), Positives = 65/81 (79%)
Query: 6 EPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYT 65
+PL V RV+G+V+D FN S+ + VTY ++V NG +L PS V NKPRV+IGGED+R+ YT
Sbjct: 7 DPLIVSRVVGDVLDPFNRSITLKVTYGQREVTNGLDLRPSQVQNKPRVEIGGEDLRNFYT 66
Query: 66 LIMTDPDAPSPSDPHLREHLH 86
L+M DPD PSPS+PHLRE+LH
Sbjct: 67 LVMVDPDVPSPSNPHLREYLH 87
>CEN_ARATH (Q9ZNV5) CENTRORADIALIS-like protein
Length = 175
Score = 111 bits (277), Expect = 4e-25
Identities = 54/82 (65%), Positives = 65/82 (78%), Gaps = 1/82 (1%)
Query: 6 EPLSVGRVIGEVVDIFNPSVRMNVTY-STKQVANGHELMPSIVMNKPRVDIGGEDMRSAY 64
+PL VGRVIG+VVD +V+M VTY S KQV NGHEL PS+V KP+V++ G DMRS +
Sbjct: 7 DPLMVGRVIGDVVDNCLQAVKMTVTYNSDKQVYNGHELFPSVVTYKPKVEVHGGDMRSFF 66
Query: 65 TLIMTDPDAPSPSDPHLREHLH 86
TL+MTDPD P PSDP+LREHLH
Sbjct: 67 TLVMTDPDVPGPSDPYLREHLH 88
>CEN_ANTMA (Q41261) CENTRORADIALIS protein
Length = 181
Score = 108 bits (270), Expect = 2e-24
Identities = 53/85 (62%), Positives = 64/85 (74%), Gaps = 4/85 (4%)
Query: 6 EPLSVGRVIGEVVDIFNPSVRMNVTY----STKQVANGHELMPSIVMNKPRVDIGGEDMR 61
+PL +GRVIG+VVD F +V+M+V Y S K V NGHEL PS V + PRV++ G DMR
Sbjct: 8 DPLVIGRVIGDVVDHFTSTVKMSVIYNSNNSIKHVYNGHELFPSAVTSTPRVEVHGGDMR 67
Query: 62 SAYTLIMTDPDAPSPSDPHLREHLH 86
S +TLIMTDPD P PSDP+LREHLH
Sbjct: 68 SFFTLIMTDPDVPGPSDPYLREHLH 92
>TSF_ARATH (Q9S7R5) TWIN SISTER of FT protein (TFL1 like protein)
Length = 175
Score = 101 bits (252), Expect = 3e-22
Identities = 46/81 (56%), Positives = 61/81 (74%)
Query: 6 EPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYT 65
+PL VG V+G+V+D F V + VTY ++V NG +L PS V+NKP V+IGG+D R+ YT
Sbjct: 7 DPLVVGSVVGDVLDPFTRLVSLKVTYGHREVTNGLDLRPSQVLNKPIVEIGGDDFRNFYT 66
Query: 66 LIMTDPDAPSPSDPHLREHLH 86
L+M DPD PSPS+PH RE+LH
Sbjct: 67 LVMVDPDVPSPSNPHQREYLH 87
>MFT_ARATH (Q9XFK7) MOTHER of FT and TF1 protein
Length = 173
Score = 93.6 bits (231), Expect = 8e-20
Identities = 43/86 (50%), Positives = 62/86 (72%), Gaps = 1/86 (1%)
Query: 1 MSRPLEPLSVGRVIGEVVDIFNPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDM 60
M+ ++PL VGRVIG+V+D+F P+ M+V + K + NG E+ PS +N P+V+I G
Sbjct: 1 MAASVDPLVVGRVIGDVLDMFIPTANMSVYFGPKHITNGCEIKPSTAVNPPKVNISGHS- 59
Query: 61 RSAYTLIMTDPDAPSPSDPHLREHLH 86
YTL+MTDPDAPSPS+P++RE +H
Sbjct: 60 DELYTLVMTDPDAPSPSEPNMREWVH 85
>OV16_ONCVO (P31729) OV-16 antigen precursor
Length = 197
Score = 52.0 bits (123), Expect = 3e-07
Identities = 30/74 (40%), Positives = 43/74 (57%), Gaps = 2/74 (2%)
Query: 15 GEVVDIFN--PSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYTLIMTDPD 72
G V D+ + P+ +NV+Y+ V G+EL P+ V N+P + + YTL+MTDPD
Sbjct: 41 GIVPDVVSTAPTKLVNVSYNNLTVNLGNELTPTQVKNQPTKVSWDAEPGALYTLVMTDPD 100
Query: 73 APSPSDPHLREHLH 86
APS +P RE H
Sbjct: 101 APSRKNPVFREWHH 114
>D1_ONCVO (P54186) D1 protein (Fragment)
Length = 152
Score = 52.0 bits (123), Expect = 3e-07
Identities = 27/65 (41%), Positives = 39/65 (59%)
Query: 22 NPSVRMNVTYSTKQVANGHELMPSIVMNKPRVDIGGEDMRSAYTLIMTDPDAPSPSDPHL 81
+P+ +NV+Y+ V G+EL P+ V N+P + + YTL+MTDPDAPS +P
Sbjct: 5 SPTKLVNVSYNNLTVNLGNELTPTQVKNQPTKVSWDAEPGALYTLVMTDPDAPSRKNPVF 64
Query: 82 REHLH 86
RE H
Sbjct: 65 REWHH 69
>PEBP_HUMAN (P30086) Phosphatidylethanolamine-binding protein
(PEBP) (Prostatic binding protein) (HCNPpp)
(Neuropolypeptide h3) (Raf kinase inhibitor protein)
(RKIP) [Contains: Hippocampal cholinergic
neurostimulating peptide (HCNP)]
Length = 186
Score = 47.0 bits (110), Expect = 8e-06
Identities = 28/66 (42%), Positives = 36/66 (54%), Gaps = 2/66 (3%)
Query: 23 PSVRMNVTYSTKQVAN-GHELMPSIVMNKPR-VDIGGEDMRSAYTLIMTDPDAPSPSDPH 80
P ++VTY+ V G L P+ V N+P + G D YTL++TDPDAPS DP
Sbjct: 20 PQHPLHVTYAGAAVDELGKVLTPTQVKNRPTSISWDGLDSGKLYTLVLTDPDAPSRKDPK 79
Query: 81 LREHLH 86
RE H
Sbjct: 80 YREWHH 85
>PEBP_MOUSE (P70296) Phosphatidylethanolamine-binding protein
(PEBP) (HCNPpp) [Contains: Hippocampal cholinergic
neurostimulating peptide (HCNP)]
Length = 186
Score = 46.6 bits (109), Expect = 1e-05
Identities = 28/66 (42%), Positives = 35/66 (52%), Gaps = 2/66 (3%)
Query: 23 PSVRMNVTYSTKQVAN-GHELMPSIVMNKPR-VDIGGEDMRSAYTLIMTDPDAPSPSDPH 80
P + V Y+ V G L P+ VMN+P + G D YTL++TDPDAPS DP
Sbjct: 20 PQHALRVDYAGVTVDELGKVLTPTQVMNRPSSISWDGLDPGKLYTLVLTDPDAPSRKDPK 79
Query: 81 LREHLH 86
RE H
Sbjct: 80 FREWHH 85
>PEBP_RAT (P31044) Phosphatidylethanolamine-binding protein (PEBP)
(HCNPpp) (23 kDa morphine-binding protein) (P23K)
[Contains: Hippocampal cholinergic neurostimulating
peptide (HCNP)]
Length = 186
Score = 46.2 bits (108), Expect = 1e-05
Identities = 24/49 (48%), Positives = 29/49 (58%), Gaps = 1/49 (2%)
Query: 39 GHELMPSIVMNKPR-VDIGGEDMRSAYTLIMTDPDAPSPSDPHLREHLH 86
G L P+ VMN+P + G D YTL++TDPDAPS DP RE H
Sbjct: 37 GKVLTPTQVMNRPSSISWDGLDPGKLYTLVLTDPDAPSRKDPKFREWHH 85
>PEBP_MACFA (P48737) Phosphatidylethanolamine-binding protein
(PEBP) (HCNPpp) [Contains: Hippocampal cholinergic
neurostimulating peptide (HCNP)]
Length = 186
Score = 45.8 bits (107), Expect = 2e-05
Identities = 27/66 (40%), Positives = 36/66 (53%), Gaps = 2/66 (3%)
Query: 23 PSVRMNVTYSTKQVAN-GHELMPSIVMNKPR-VDIGGEDMRSAYTLIMTDPDAPSPSDPH 80
P ++VTY+ + G L P+ V N+P + G D YTL++TDPDAPS DP
Sbjct: 20 PQHPLHVTYAGAALDELGKVLTPTQVKNRPTSISWDGLDSGKLYTLVLTDPDAPSRKDPK 79
Query: 81 LREHLH 86
RE H
Sbjct: 80 YREWHH 85
>PEBP_BOVIN (P13696) Phosphatidylethanolamine-binding protein
(PEBP) (HCNPpp) (Basic cytosolic 21 kDa protein)
[Contains: Hippocampal cholinergic neurostimulating
peptide (HCNP)]
Length = 186
Score = 44.3 bits (103), Expect = 5e-05
Identities = 27/66 (40%), Positives = 34/66 (50%), Gaps = 2/66 (3%)
Query: 23 PSVRMNVTYSTKQVAN-GHELMPSIVMNKPR-VDIGGEDMRSAYTLIMTDPDAPSPSDPH 80
P + V Y +V G L P+ V N+P + G D YTL++TDPDAPS DP
Sbjct: 20 PQHPLQVKYGGAEVDELGKVLTPTQVKNRPTSITWDGLDPGKLYTLVLTDPDAPSRKDPK 79
Query: 81 LREHLH 86
RE H
Sbjct: 80 YREWHH 85
>D3_ONCVO (P54188) D3 protein (Fragment)
Length = 134
Score = 43.9 bits (102), Expect = 7e-05
Identities = 22/48 (45%), Positives = 29/48 (59%)
Query: 39 GHELMPSIVMNKPRVDIGGEDMRSAYTLIMTDPDAPSPSDPHLREHLH 86
G+EL P+ V N+P + + YTL+MTDPDAPS +P RE H
Sbjct: 4 GNELTPTQVKNQPTKVSWDAEPGALYTLVMTDPDAPSRKNPVFREWHH 51
>PEB2_MOUSE (Q8VIN1) Phosphatidylethanolamine-binding protein-2
(PEBP-2)
Length = 187
Score = 43.5 bits (101), Expect = 9e-05
Identities = 27/66 (40%), Positives = 35/66 (52%), Gaps = 2/66 (3%)
Query: 23 PSVRMNVTYSTKQVAN-GHELMPSIVMNKP-RVDIGGEDMRSAYTLIMTDPDAPSPSDPH 80
P + VTY+ +V G L P+ V ++P + G D YTLI+TDPDAPS P
Sbjct: 21 PQHLLRVTYTEAEVEELGQVLTPTQVKHRPGSISWDGLDPGKLYTLILTDPDAPSRKKPV 80
Query: 81 LREHLH 86
RE H
Sbjct: 81 YREWHH 86
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.141 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,118,809
Number of Sequences: 164201
Number of extensions: 450911
Number of successful extensions: 1255
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1213
Number of HSP's gapped (non-prelim): 37
length of query: 97
length of database: 59,974,054
effective HSP length: 73
effective length of query: 24
effective length of database: 47,987,381
effective search space: 1151697144
effective search space used: 1151697144
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC146807.7


